Danni Chen
Guangdong University of Technology, Guangzhou, China
Fusion of Various Optimization Based Feature Smoothing Methods for Wearable and Non-invasive Blood Glucose Estimation
Mar 06, 2025



Abstract:Recently, the wearable and non-invasive blood glucose estimation approach has been proposed. However, due to the unreliability of the acquisition device, the presence of the noise and the variations of the acquisition environments, the obtained features and the reference blood glucose values are highly unreliable. To address this issue, this paper proposes a polynomial fitting approach to smooth the obtained features or the reference blood glucose values. First, the blood glucose values are estimated based on the individual optimization approaches. Second, the absolute difference values between the estimated blood glucose values and the actual blood glucose values based on each optimization approach are computed. Third, these absolute difference values for each optimization approach are sorted in the ascending order. Fourth, for each sorted blood glucose value, the optimization method corresponding to the minimum absolute difference value is selected. Fifth, the accumulate probability of each selected optimization method is computed. If the accumulate probability of any selected optimization method at a point is greater than a threshold value, then the accumulate probabilities of these three selected optimization methods at that point are reset to zero. A range of the sorted blood glucose values are defined as that with the corresponding boundaries points being the previous reset point and this reset point. Hence, after performing the above procedures for all the sorted reference blood glucose values in the validation set, the regions of the sorted reference blood glucose values and the corresponding optimization methods in these regions are determined. The computer numerical simulation results show that our proposed method yields the mean absolute relative deviation (MARD) at 0.0930 and the percentage of the test data falling in the zone A of the Clarke error grid at 94.1176%.
* This version corrects several typos
SOFFLFM: Super-resolution optical fluctuation Fourier light-field microscopy
Aug 26, 2022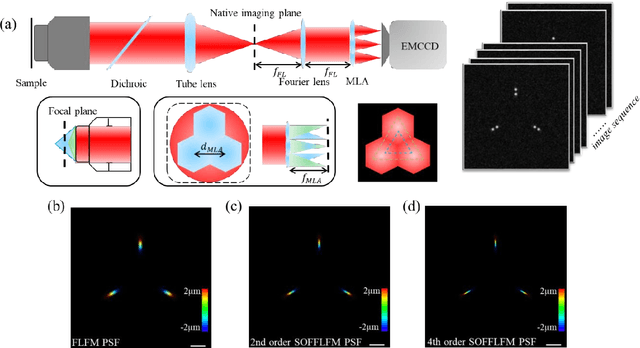

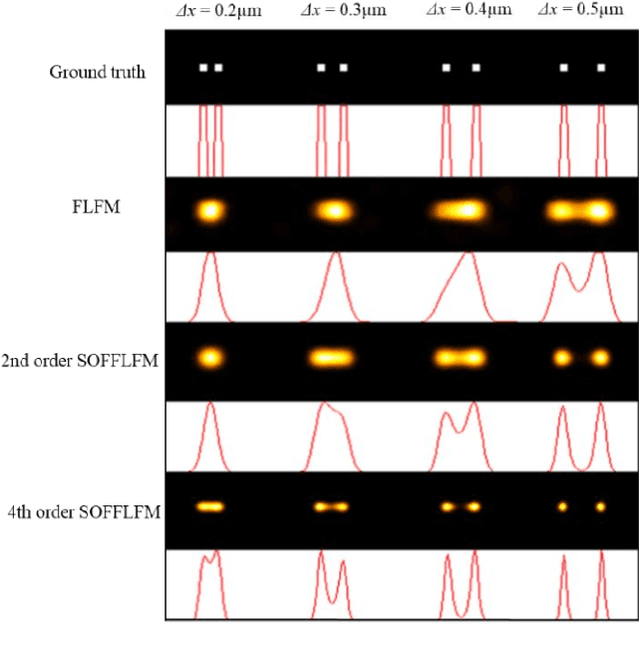
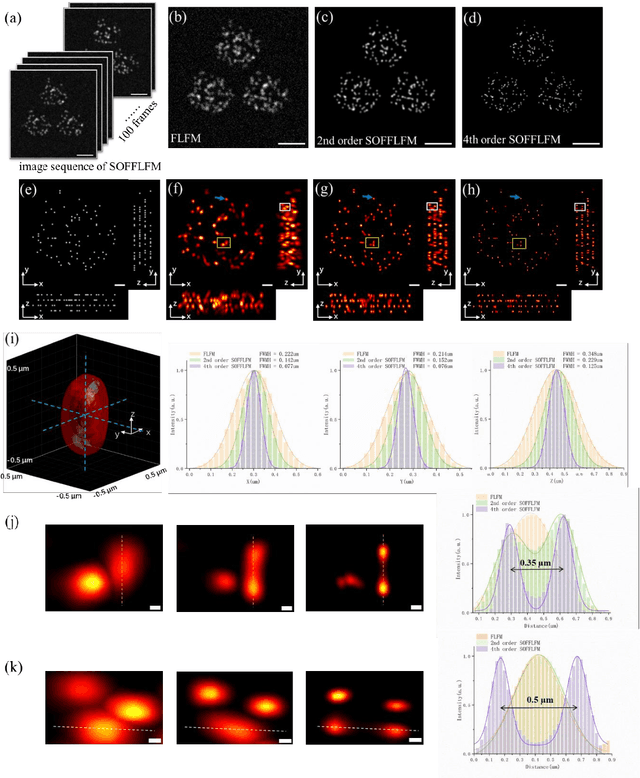
Abstract:Fourier light-field microscopy (FLFM) uses a micro-lens array (MLA) to segment the Fourier Plane of the microscopic objective lens to generate multiple two-dimensional perspective views, thereby reconstructing the three-dimensional(3D) structure of the sample using 3D deconvolution calculation without scanning. However, the resolution of FLFM is still limited by diffraction, and furthermore, dependent on the aperture division. In order to improve its resolution, a Super-resolution optical fluctuation Fourier light field microscopy (SOFFLFM) was proposed here, in which the Sofi method with ability of super-resolution was introduced into FLFM. SOFFLFM uses higher-order cumulants statistical analysis on an image sequence collected by FLFM, and then carries out 3D deconvolution calculation to reconstruct the 3D structure of the sample. Theoretical basis of SOFFLFM on improving resolution was explained and then verified with simulations. Simulation results demonstrated that SOFFLFM improved lateral and axial resolution by more than sqrt(2) and 2 times in the 2nd and 4th order accumulations, compared with that of FLFM.
Multi-institutional Validation of Two-Streamed Deep Learning Method for Automated Delineation of Esophageal Gross Tumor Volume using planning-CT and FDG-PETCT
Oct 11, 2021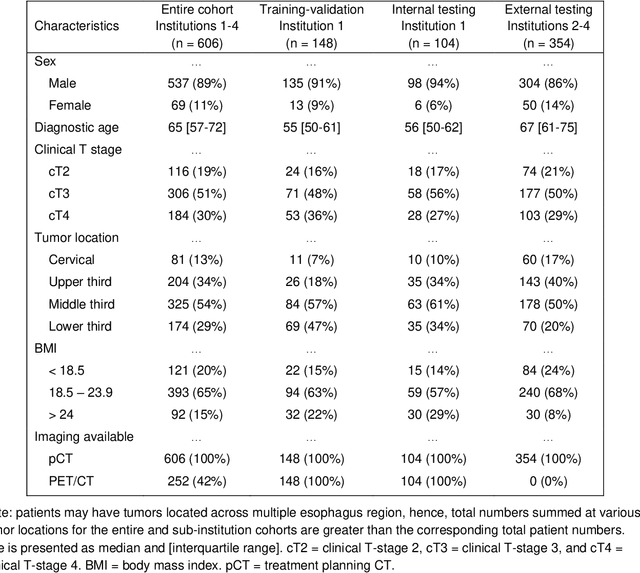
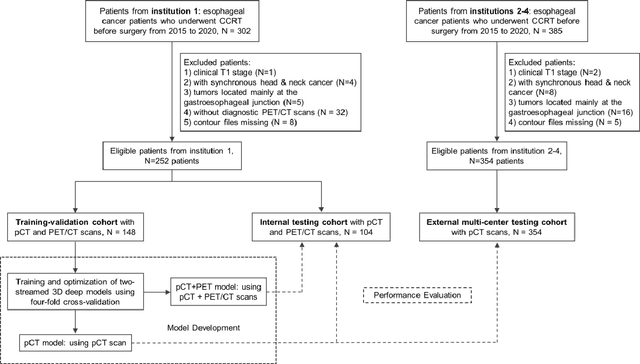
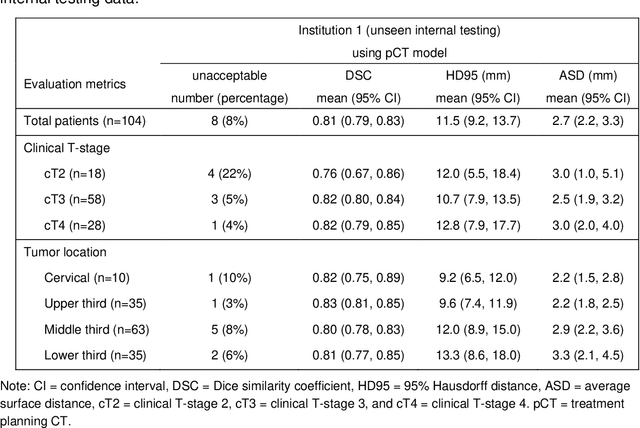
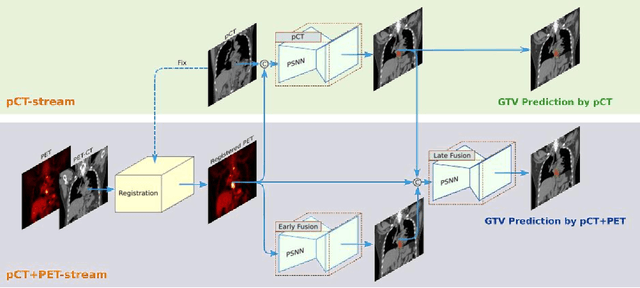
Abstract:Background: The current clinical workflow for esophageal gross tumor volume (GTV) contouring relies on manual delineation of high labor-costs and interuser variability. Purpose: To validate the clinical applicability of a deep learning (DL) multi-modality esophageal GTV contouring model, developed at 1 institution whereas tested at multiple ones. Methods and Materials: We collected 606 esophageal cancer patients from four institutions. 252 institution-1 patients had a treatment planning-CT (pCT) and a pair of diagnostic FDG-PETCT; 354 patients from other 3 institutions had only pCT. A two-streamed DL model for GTV segmentation was developed using pCT and PETCT scans of a 148 patient institution-1 subset. This built model had the flexibility of segmenting GTVs via only pCT or pCT+PETCT combined. For independent evaluation, the rest 104 institution-1 patients behaved as unseen internal testing, and 354 institutions 2-4 patients were used for external testing. We evaluated manual revision degrees by human experts to assess the contour-editing effort. The performance of the deep model was compared against 4 radiation oncologists in a multiuser study with 20 random external patients. Contouring accuracy and time were recorded for the pre-and post-DL assisted delineation process. Results: Our model achieved high segmentation accuracy in internal testing (mean Dice score: 0.81 using pCT and 0.83 using pCT+PET) and generalized well to external evaluation (mean DSC: 0.80). Expert assessment showed that the predicted contours of 88% patients need only minor or no revision. In multi-user evaluation, with the assistance of a deep model, inter-observer variation and required contouring time were reduced by 37.6% and 48.0%, respectively. Conclusions: Deep learning predicted GTV contours were in close agreement with the ground truth and could be adopted clinically with mostly minor or no changes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge