Danilo Dessì
Research Knowledge Graphs: the Shifting Paradigm of Scholarly Information Representation
Jun 08, 2025Abstract:Sharing and reusing research artifacts, such as datasets, publications, or methods is a fundamental part of scientific activity, where heterogeneity of resources and metadata and the common practice of capturing information in unstructured publications pose crucial challenges. Reproducibility of research and finding state-of-the-art methods or data have become increasingly challenging. In this context, the concept of Research Knowledge Graphs (RKGs) has emerged, aiming at providing an easy to use and machine-actionable representation of research artifacts and their relations. That is facilitated through the use of established principles for data representation, the consistent adoption of globally unique persistent identifiers and the reuse and linking of vocabularies and data. This paper provides the first conceptualisation of the RKG vision, a categorisation of in-use RKGs together with a description of RKG building blocks and principles. We also survey real-world RKG implementations differing with respect to scale, schema, data, used vocabulary, and reliability of the contained data. We also characterise different RKG construction methodologies and provide a forward-looking perspective on the diverse applications, opportunities, and challenges associated with the RKG vision.
Triplètoile: Extraction of Knowledge from Microblogging Text
Aug 27, 2024



Abstract:Numerous methods and pipelines have recently emerged for the automatic extraction of knowledge graphs from documents such as scientific publications and patents. However, adapting these methods to incorporate alternative text sources like micro-blogging posts and news has proven challenging as they struggle to model open-domain entities and relations, typically found in these sources. In this paper, we propose an enhanced information extraction pipeline tailored to the extraction of a knowledge graph comprising open-domain entities from micro-blogging posts on social media platforms. Our pipeline leverages dependency parsing and classifies entity relations in an unsupervised manner through hierarchical clustering over word embeddings. We provide a use case on extracting semantic triples from a corpus of 100 thousand tweets about digital transformation and publicly release the generated knowledge graph. On the same dataset, we conduct two experimental evaluations, showing that the system produces triples with precision over 95% and outperforms similar pipelines of around 5% in terms of precision, while generating a comparatively higher number of triples.
* 42 pages, 6 figures
Generating Knowledge Graphs by Employing Natural Language Processing and Machine Learning Techniques within the Scholarly Domain
Oct 28, 2020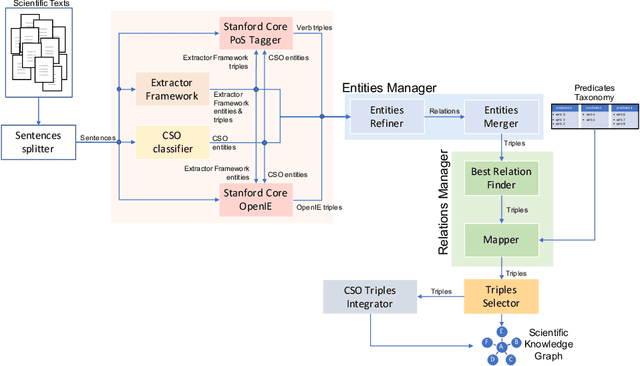
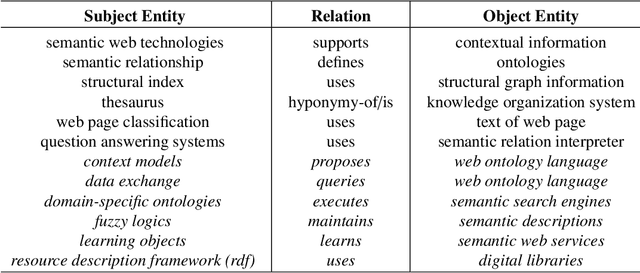
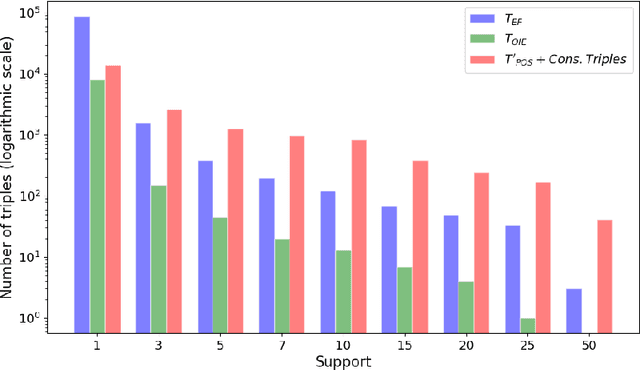
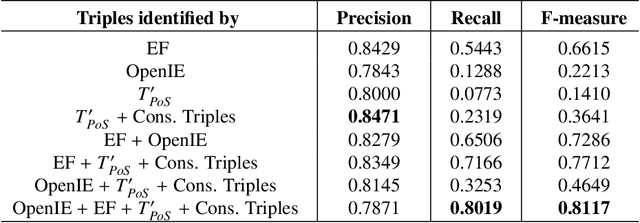
Abstract:The continuous growth of scientific literature brings innovations and, at the same time, raises new challenges. One of them is related to the fact that its analysis has become difficult due to the high volume of published papers for which manual effort for annotations and management is required. Novel technological infrastructures are needed to help researchers, research policy makers, and companies to time-efficiently browse, analyse, and forecast scientific research. Knowledge graphs i.e., large networks of entities and relationships, have proved to be effective solution in this space. Scientific knowledge graphs focus on the scholarly domain and typically contain metadata describing research publications such as authors, venues, organizations, research topics, and citations. However, the current generation of knowledge graphs lacks of an explicit representation of the knowledge presented in the research papers. As such, in this paper, we present a new architecture that takes advantage of Natural Language Processing and Machine Learning methods for extracting entities and relationships from research publications and integrates them in a large-scale knowledge graph. Within this research work, we i) tackle the challenge of knowledge extraction by employing several state-of-the-art Natural Language Processing and Text Mining tools, ii) describe an approach for integrating entities and relationships generated by these tools, iii) show the advantage of such an hybrid system over alternative approaches, and vi) as a chosen use case, we generated a scientific knowledge graph including 109,105 triples, extracted from 26,827 abstracts of papers within the Semantic Web domain. As our approach is general and can be applied to any domain, we expect that it can facilitate the management, analysis, dissemination, and processing of scientific knowledge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge