Confidence Raymond
Training Beyond Convergence: Grokking nnU-Net for Glioma Segmentation in Sub-Saharan MRI
Jan 30, 2026Abstract:Gliomas are placing an increasingly clinical burden on Sub-Saharan Africa (SSA). In the region, the median survival for patients remains under two years, and access to diagnostic imaging is extremely limited. These constraints highlight an urgent need for automated tools that can extract the maximum possible information from each available scan, tools that are specifically trained on local data, rather than adapted from high-income settings where conditions are vastly different. We utilize the Brain Tumor Segmentation (BraTS) Africa 2025 Challenge dataset, an expert annotated collection of glioma MRIs. Our objectives are: (i) establish a strong baseline with nnUNet on this dataset, and (ii) explore whether the celebrated "grokking" phenomenon an abrupt, late training jump from memorization to superior generalization can be triggered to push performance without extra labels. We evaluate two training regimes. The first is a fast, budget-conscious approach that limits optimization to just a few epochs, reflecting the constrained GPU resources typically available in African institutions. Despite this limitation, nnUNet achieves strong Dice scores: 92.3% for whole tumor (WH), 86.6% for tumor core (TC), and 86.3% for enhancing tumor (ET). The second regime extends training well beyond the point of convergence, aiming to trigger a grokking-driven performance leap. With this approach, we were able to achieve grokking and enhanced our results to higher Dice scores: 92.2% for whole tumor (WH), 90.1% for tumor core (TC), and 90.2% for enhancing tumor (ET).
Resource-Efficient Glioma Segmentation on Sub-Saharan MRI
Sep 11, 2025Abstract:Gliomas are the most prevalent type of primary brain tumors, and their accurate segmentation from MRI is critical for diagnosis, treatment planning, and longitudinal monitoring. However, the scarcity of high-quality annotated imaging data in Sub-Saharan Africa (SSA) poses a significant challenge for deploying advanced segmentation models in clinical workflows. This study introduces a robust and computationally efficient deep learning framework tailored for resource-constrained settings. We leveraged a 3D Attention UNet architecture augmented with residual blocks and enhanced through transfer learning from pre-trained weights on the BraTS 2021 dataset. Our model was evaluated on 95 MRI cases from the BraTS-Africa dataset, a benchmark for glioma segmentation in SSA MRI data. Despite the limited data quality and quantity, our approach achieved Dice scores of 0.76 for the Enhancing Tumor (ET), 0.80 for Necrotic and Non-Enhancing Tumor Core (NETC), and 0.85 for Surrounding Non-Functional Hemisphere (SNFH). These results demonstrate the generalizability of the proposed model and its potential to support clinical decision making in low-resource settings. The compact architecture, approximately 90 MB, and sub-minute per-volume inference time on consumer-grade hardware further underscore its practicality for deployment in SSA health systems. This work contributes toward closing the gap in equitable AI for global health by empowering underserved regions with high-performing and accessible medical imaging solutions.
Towards Trustworthy Breast Tumor Segmentation in Ultrasound using Monte Carlo Dropout and Deep Ensembles for Epistemic Uncertainty Estimation
Aug 25, 2025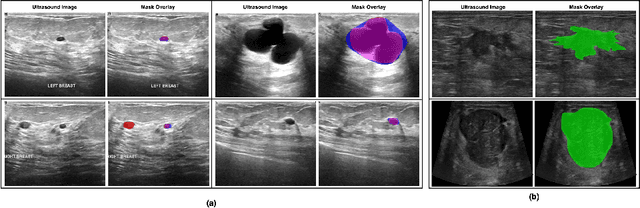
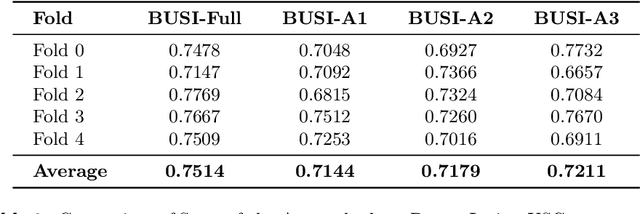
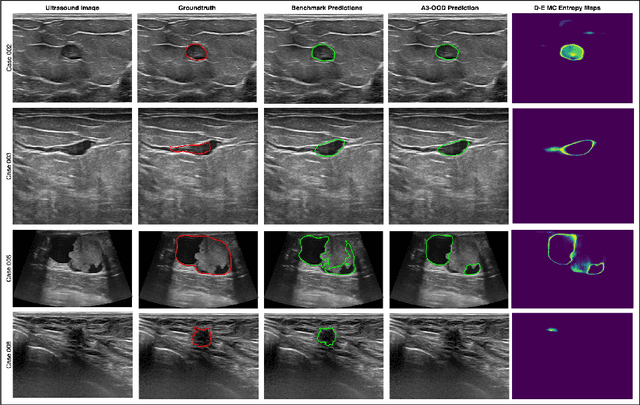
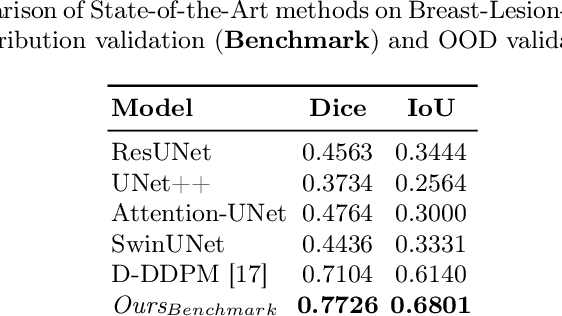
Abstract:Automated segmentation of BUS images is important for precise lesion delineation and tumor characterization, but is challenged by inherent artifacts and dataset inconsistencies. In this work, we evaluate the use of a modified Residual Encoder U-Net for breast ultrasound segmentation, with a focus on uncertainty quantification. We identify and correct for data duplication in the BUSI dataset, and use a deduplicated subset for more reliable estimates of generalization performance. Epistemic uncertainty is quantified using Monte Carlo dropout, deep ensembles, and their combination. Models are benchmarked on both in-distribution and out-of-distribution datasets to demonstrate how they generalize to unseen cross-domain data. Our approach achieves state-of-the-art segmentation accuracy on the Breast-Lesion-USG dataset with in-distribution validation, and provides calibrated uncertainty estimates that effectively signal regions of low model confidence. Performance declines and increased uncertainty observed in out-of-distribution evaluation highlight the persistent challenge of domain shift in medical imaging, and the importance of integrated uncertainty modeling for trustworthy clinical deployment. \footnote{Code available at: https://github.com/toufiqmusah/nn-uncertainty.git}
Parameter-efficient Fine-tuning for improved Convolutional Baseline for Brain Tumor Segmentation in Sub-Saharan Africa Adult Glioma Dataset
Dec 18, 2024



Abstract:Automating brain tumor segmentation using deep learning methods is an ongoing challenge in medical imaging. Multiple lingering issues exist including domain-shift and applications in low-resource settings which brings a unique set of challenges including scarcity of data. As a step towards solving these specific problems, we propose Convolutional adapter-inspired Parameter-efficient Fine-tuning (PEFT) of MedNeXt architecture. To validate our idea, we show our method performs comparable to full fine-tuning with the added benefit of reduced training compute using BraTS-2021 as pre-training dataset and BraTS-Africa as the fine-tuning dataset. BraTS-Africa consists of a small dataset (60 train / 35 validation) from the Sub-Saharan African population with marked shift in the MRI quality compared to BraTS-2021 (1251 train samples). We first show that models trained on BraTS-2021 dataset do not generalize well to BraTS-Africa as shown by 20% reduction in mean dice on BraTS-Africa validation samples. Then, we show that PEFT can leverage both the BraTS-2021 and BraTS-Africa dataset to obtain mean dice of 0.8 compared to 0.72 when trained only on BraTS-Africa. Finally, We show that PEFT (0.80 mean dice) results in comparable performance to full fine-tuning (0.77 mean dice) which may show PEFT to be better on average but the boxplots show that full finetuning results is much lesser variance in performance. Nevertheless, on disaggregation of the dice metrics, we find that the model has tendency to oversegment as shown by high specificity (0.99) compared to relatively low sensitivity(0.75). The source code is available at https://github.com/CAMERA-MRI/SPARK2024/tree/main/PEFT_MedNeXt
Bridging the Gap: Generalising State-of-the-Art U-Net Models to Sub-Saharan African Populations
Dec 19, 2023Abstract:A critical challenge for tumour segmentation models is the ability to adapt to diverse clinical settings, particularly when applied to poor-quality neuroimaging data. The uncertainty surrounding this adaptation stems from the lack of representative datasets, leaving top-performing models without exposure to common artifacts found in MRI data throughout Sub-Saharan Africa (SSA). We replicated a framework that secured the 2nd position in the 2022 BraTS competition to investigate the impact of dataset composition on model performance and pursued four distinct approaches through training a model with: 1) BraTS-Africa data only (train_SSA, N=60), 2) BraTS-Adult Glioma data only (train_GLI, N=1251), 3) both datasets together (train_ALL, N=1311), and 4) through further training the train_GLI model with BraTS-Africa data (train_ftSSA). Notably, training on a smaller low-quality dataset alone (train_SSA) yielded subpar results, and training on a larger high-quality dataset alone (train_GLI) struggled to delineate oedematous tissue in the low-quality validation set. The most promising approach (train_ftSSA) involved pre-training a model on high-quality neuroimages and then fine-tuning it on the smaller, low-quality dataset. This approach outperformed the others, ranking second in the MICCAI BraTS Africa global challenge external testing phase. These findings underscore the significance of larger sample sizes and broad exposure to data in improving segmentation performance. Furthermore, we demonstrated that there is potential for improving such models by fine-tuning them with a wider range of data locally.
The Brain Tumor Segmentation Challenge 2023: Glioma Segmentation in Sub-Saharan Africa Patient Population
May 30, 2023

Abstract:Gliomas are the most common type of primary brain tumors. Although gliomas are relatively rare, they are among the deadliest types of cancer, with a survival rate of less than 2 years after diagnosis. Gliomas are challenging to diagnose, hard to treat and inherently resistant to conventional therapy. Years of extensive research to improve diagnosis and treatment of gliomas have decreased mortality rates across the Global North, while chances of survival among individuals in low- and middle-income countries (LMICs) remain unchanged and are significantly worse in Sub-Saharan Africa (SSA) populations. Long-term survival with glioma is associated with the identification of appropriate pathological features on brain MRI and confirmation by histopathology. Since 2012, the Brain Tumor Segmentation (BraTS) Challenge have evaluated state-of-the-art machine learning methods to detect, characterize, and classify gliomas. However, it is unclear if the state-of-the-art methods can be widely implemented in SSA given the extensive use of lower-quality MRI technology, which produces poor image contrast and resolution and more importantly, the propensity for late presentation of disease at advanced stages as well as the unique characteristics of gliomas in SSA (i.e., suspected higher rates of gliomatosis cerebri). Thus, the BraTS-Africa Challenge provides a unique opportunity to include brain MRI glioma cases from SSA in global efforts through the BraTS Challenge to develop and evaluate computer-aided-diagnostic (CAD) methods for the detection and characterization of glioma in resource-limited settings, where the potential for CAD tools to transform healthcare are more likely.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge