Charidimos Tsagkas
Benchmarking and Explaining Deep Learning Cortical Lesion MRI Segmentation in Multiple Sclerosis
Jul 16, 2025Abstract:Cortical lesions (CLs) have emerged as valuable biomarkers in multiple sclerosis (MS), offering high diagnostic specificity and prognostic relevance. However, their routine clinical integration remains limited due to subtle magnetic resonance imaging (MRI) appearance, challenges in expert annotation, and a lack of standardized automated methods. We propose a comprehensive multi-centric benchmark of CL detection and segmentation in MRI. A total of 656 MRI scans, including clinical trial and research data from four institutions, were acquired at 3T and 7T using MP2RAGE and MPRAGE sequences with expert-consensus annotations. We rely on the self-configuring nnU-Net framework, designed for medical imaging segmentation, and propose adaptations tailored to the improved CL detection. We evaluated model generalization through out-of-distribution testing, demonstrating strong lesion detection capabilities with an F1-score of 0.64 and 0.5 in and out of the domain, respectively. We also analyze internal model features and model errors for a better understanding of AI decision-making. Our study examines how data variability, lesion ambiguity, and protocol differences impact model performance, offering future recommendations to address these barriers to clinical adoption. To reinforce the reproducibility, the implementation and models will be publicly accessible and ready to use at https://github.com/Medical-Image-Analysis-Laboratory/ and https://doi.org/10.5281/zenodo.15911797.
Monitoring morphometric drift in lifelong learning segmentation of the spinal cord
May 02, 2025Abstract:Morphometric measures derived from spinal cord segmentations can serve as diagnostic and prognostic biomarkers in neurological diseases and injuries affecting the spinal cord. While robust, automatic segmentation methods to a wide variety of contrasts and pathologies have been developed over the past few years, whether their predictions are stable as the model is updated using new datasets has not been assessed. This is particularly important for deriving normative values from healthy participants. In this study, we present a spinal cord segmentation model trained on a multisite $(n=75)$ dataset, including 9 different MRI contrasts and several spinal cord pathologies. We also introduce a lifelong learning framework to automatically monitor the morphometric drift as the model is updated using additional datasets. The framework is triggered by an automatic GitHub Actions workflow every time a new model is created, recording the morphometric values derived from the model's predictions over time. As a real-world application of the proposed framework, we employed the spinal cord segmentation model to update a recently-introduced normative database of healthy participants containing commonly used measures of spinal cord morphometry. Results showed that: (i) our model outperforms previous versions and pathology-specific models on challenging lumbar spinal cord cases, achieving an average Dice score of $0.95 \pm 0.03$; (ii) the automatic workflow for monitoring morphometric drift provides a quick feedback loop for developing future segmentation models; and (iii) the scaling factor required to update the database of morphometric measures is nearly constant among slices across the given vertebral levels, showing minimum drift between the current and previous versions of the model monitored by the framework. The model is freely available in Spinal Cord Toolbox v7.0.
Towards contrast-agnostic soft segmentation of the spinal cord
Oct 23, 2023



Abstract:Spinal cord segmentation is clinically relevant and is notably used to compute spinal cord cross-sectional area (CSA) for the diagnosis and monitoring of cord compression or neurodegenerative diseases such as multiple sclerosis. While several semi and automatic methods exist, one key limitation remains: the segmentation depends on the MRI contrast, resulting in different CSA across contrasts. This is partly due to the varying appearance of the boundary between the spinal cord and the cerebrospinal fluid that depends on the sequence and acquisition parameters. This contrast-sensitive CSA adds variability in multi-center studies where protocols can vary, reducing the sensitivity to detect subtle atrophies. Moreover, existing methods enhance the CSA variability by training one model per contrast, while also producing binary masks that do not account for partial volume effects. In this work, we present a deep learning-based method that produces soft segmentations of the spinal cord. Using the Spine Generic Public Database of healthy participants ($\text{n}=267$; $\text{contrasts}=6$), we first generated participant-wise soft ground truth (GT) by averaging the binary segmentations across all 6 contrasts. These soft GT, along with a regression-based loss function, were then used to train a UNet model for spinal cord segmentation. We evaluated our model against state-of-the-art methods and performed ablation studies involving different GT mask types, loss functions, and contrast-specific models. Our results show that using the soft average segmentations along with a regression loss function reduces CSA variability ($p < 0.05$, Wilcoxon signed-rank test). The proposed spinal cord segmentation model generalizes better than the state-of-the-art contrast-specific methods amongst unseen datasets, vendors, contrasts, and pathologies (compression, lesions), while accounting for partial volume effects.
Spinal Cord Gray Matter-White Matter Segmentation on Magnetic Resonance AMIRA Images with MD-GRU
Aug 07, 2018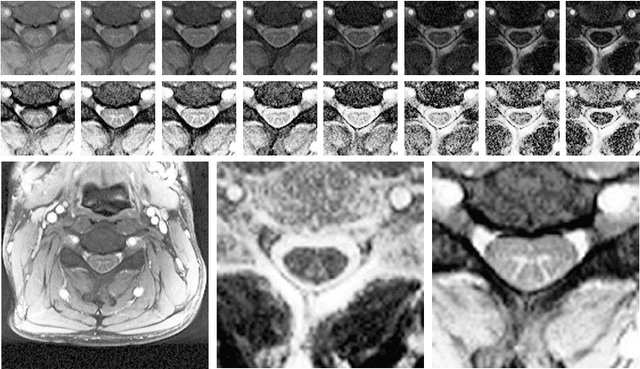
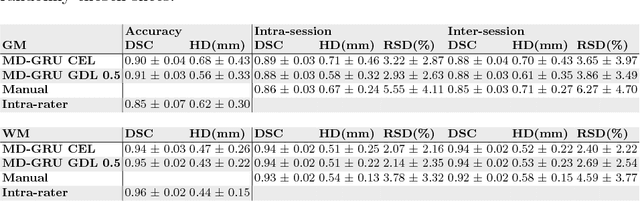
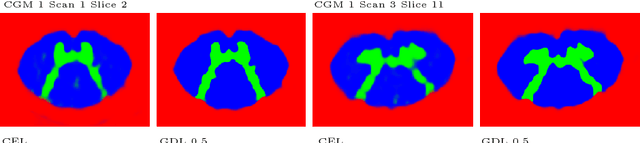
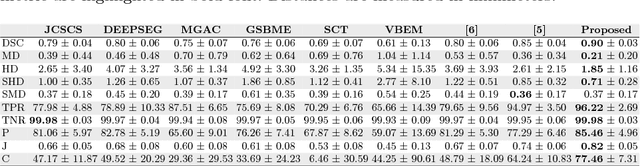
Abstract:The small butterfly shaped structure of spinal cord (SC) gray matter (GM) is challenging to image and to delinate from its surrounding white matter (WM). Segmenting GM is up to a point a trade-off between accuracy and precision. We propose a new pipeline for GM-WM magnetic resonance (MR) image acquisition and segmentation. We report superior results as compared to the ones recently reported in the SC GM segmentation challenge and show even better results using the averaged magnetization inversion recovery acquisitions (AMIRA) sequence. Scan-rescan experiments with the AMIRA sequence show high reproducibility in terms of Dice coefficient, Hausdorff distance and relative standard deviation. We use a recurrent neural network (RNN) with multi-dimensional gated recurrent units (MD-GRU) to train segmentation models on the AMIRA dataset of 855 slices. We added a generalized dice loss to the cross entropy loss that MD-GRU uses and were able to improve the results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge