Cecilia Casolo
Identifiability Challenges in Sparse Linear Ordinary Differential Equations
Jun 12, 2025Abstract:Dynamical systems modeling is a core pillar of scientific inquiry across natural and life sciences. Increasingly, dynamical system models are learned from data, rendering identifiability a paramount concept. For systems that are not identifiable from data, no guarantees can be given about their behavior under new conditions and inputs, or about possible control mechanisms to steer the system. It is known in the community that "linear ordinary differential equations (ODE) are almost surely identifiable from a single trajectory." However, this only holds for dense matrices. The sparse regime remains underexplored, despite its practical relevance with sparsity arising naturally in many biological, social, and physical systems. In this work, we address this gap by characterizing the identifiability of sparse linear ODEs. Contrary to the dense case, we show that sparse systems are unidentifiable with a positive probability in practically relevant sparsity regimes and provide lower bounds for this probability. We further study empirically how this theoretical unidentifiability manifests in state-of-the-art methods to estimate linear ODEs from data. Our results corroborate that sparse systems are also practically unidentifiable. Theoretical limitations are not resolved through inductive biases or optimization dynamics. Our findings call for rethinking what can be expected from data-driven dynamical system modeling and allows for quantitative assessments of how much to trust a learned linear ODE.
An Asymmetric Independence Model for Causal Discovery on Path Spaces
Mar 12, 2025Abstract:We develop the theory linking 'E-separation' in directed mixed graphs (DMGs) with conditional independence relations among coordinate processes in stochastic differential equations (SDEs), where causal relationships are determined by "which variables enter the governing equation of which other variables". We prove a global Markov property for cyclic SDEs, which naturally extends to partially observed cyclic SDEs, because our asymmetric independence model is closed under marginalization. We then characterize the class of graphs that encode the same set of independence relations, yielding a result analogous to the seminal 'same skeleton and v-structures' result for directed acyclic graphs (DAGs). In the fully observed case, we show that each such equivalence class of graphs has a greatest element as a parsimonious representation and develop algorithms to identify this greatest element from data. We conjecture that a greatest element also exists under partial observations, which we verify computationally for graphs with up to four nodes.
Your Assumed DAG is Wrong and Here's How To Deal With It
Feb 24, 2025Abstract:Assuming a directed acyclic graph (DAG) that represents prior knowledge of causal relationships between variables is a common starting point for cause-effect estimation. Existing literature typically invokes hypothetical domain expert knowledge or causal discovery algorithms to justify this assumption. In practice, neither may propose a single DAG with high confidence. Domain experts are hesitant to rule out dependencies with certainty or have ongoing disputes about relationships; causal discovery often relies on untestable assumptions itself or only provides an equivalence class of DAGs and is commonly sensitive to hyperparameter and threshold choices. We propose an efficient, gradient-based optimization method that provides bounds for causal queries over a collection of causal graphs -- compatible with imperfect prior knowledge -- that may still be too large for exhaustive enumeration. Our bounds achieve good coverage and sharpness for causal queries such as average treatment effects in linear and non-linear synthetic settings as well as on real-world data. Our approach aims at providing an easy-to-use and widely applicable rebuttal to the valid critique of `What if your assumed DAG is wrong?'.
Uncertainty-Aware Optimal Treatment Selection for Clinical Time Series
Oct 11, 2024Abstract:In personalized medicine, the ability to predict and optimize treatment outcomes across various time frames is essential. Additionally, the ability to select cost-effective treatments within specific budget constraints is critical. Despite recent advancements in estimating counterfactual trajectories, a direct link to optimal treatment selection based on these estimates is missing. This paper introduces a novel method integrating counterfactual estimation techniques and uncertainty quantification to recommend personalized treatment plans adhering to predefined cost constraints. Our approach is distinctive in its handling of continuous treatment variables and its incorporation of uncertainty quantification to improve prediction reliability. We validate our method using two simulated datasets, one focused on the cardiovascular system and the other on COVID-19. Our findings indicate that our method has robust performance across different counterfactual estimation baselines, showing that introducing uncertainty quantification in these settings helps the current baselines in finding more reliable and accurate treatment selection. The robustness of our method across various settings highlights its potential for broad applicability in personalized healthcare solutions.
Signature Kernel Conditional Independence Tests in Causal Discovery for Stochastic Processes
Feb 28, 2024



Abstract:Inferring the causal structure underlying stochastic dynamical systems from observational data holds great promise in domains ranging from science and health to finance. Such processes can often be accurately modeled via stochastic differential equations (SDEs), which naturally imply causal relationships via "which variables enter the differential of which other variables". In this paper, we develop a kernel-based test of conditional independence (CI) on "path-space" -- solutions to SDEs -- by leveraging recent advances in signature kernels. We demonstrate strictly superior performance of our proposed CI test compared to existing approaches on path-space. Then, we develop constraint-based causal discovery algorithms for acyclic stochastic dynamical systems (allowing for loops) that leverage temporal information to recover the entire directed graph. Assuming faithfulness and a CI oracle, our algorithm is sound and complete. We empirically verify that our developed CI test in conjunction with the causal discovery algorithm reliably outperforms baselines across a range of settings.
Learning Counterfactually Invariant Predictors
Jul 20, 2022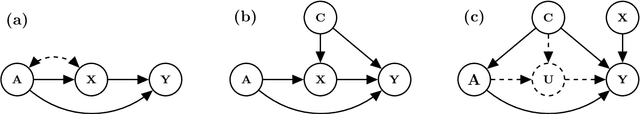
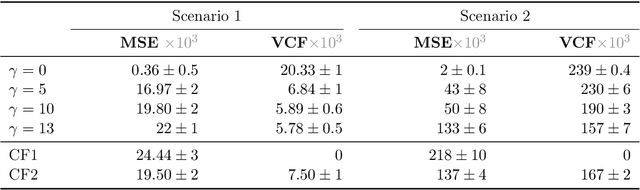

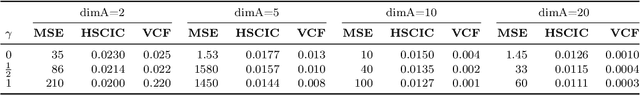
Abstract:We propose a method to learn predictors that are invariant under counterfactual changes of certain covariates. This method is useful when the prediction target is causally influenced by covariates that should not affect the predictor output. For instance, an object recognition model may be influenced by position, orientation, or scale of the object itself. We address the problem of training predictors that are explicitly counterfactually invariant to changes of such covariates. We propose a model-agnostic regularization term based on conditional kernel mean embeddings, to enforce counterfactual invariance during training. We prove the soundness of our method, which can handle mixed categorical and continuous multi-variate attributes. Empirical results on synthetic and real-world data demonstrate the efficacy of our method in a variety of settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge