Catherine R. Jutzeler
Mechanistic Learning with Guided Diffusion Models to Predict Spatio-Temporal Brain Tumor Growth
Sep 11, 2025Abstract:Predicting the spatio-temporal progression of brain tumors is essential for guiding clinical decisions in neuro-oncology. We propose a hybrid mechanistic learning framework that combines a mathematical tumor growth model with a guided denoising diffusion implicit model (DDIM) to synthesize anatomically feasible future MRIs from preceding scans. The mechanistic model, formulated as a system of ordinary differential equations, captures temporal tumor dynamics including radiotherapy effects and estimates future tumor burden. These estimates condition a gradient-guided DDIM, enabling image synthesis that aligns with both predicted growth and patient anatomy. We train our model on the BraTS adult and pediatric glioma datasets and evaluate on 60 axial slices of in-house longitudinal pediatric diffuse midline glioma (DMG) cases. Our framework generates realistic follow-up scans based on spatial similarity metrics. It also introduces tumor growth probability maps, which capture both clinically relevant extent and directionality of tumor growth as shown by 95th percentile Hausdorff Distance. The method enables biologically informed image generation in data-limited scenarios, offering generative-space-time predictions that account for mechanistic priors.
Diffusion-Based Semantic Segmentation of Lumbar Spine MRI Scans of Lower Back Pain Patients
Nov 19, 2024Abstract:This study introduces a diffusion-based framework for robust and accurate segmenton of vertebrae, intervertebral discs (IVDs), and spinal canal from Magnetic Resonance Imaging~(MRI) scans of patients with low back pain (LBP), regardless of whether the scans are T1w or T2-weighted. The results showed that SpineSegDiff achieved comparable outperformed non-diffusion state-of-the-art models in the identification of degenerated IVDs. Our findings highlight the potential of diffusion models to improve LBP diagnosis and management through precise spine MRI analysis.
Genetic prediction of quantitative traits: a machine learner's guide focused on height
Oct 06, 2023
Abstract:Machine learning and deep learning have been celebrating many successes in the application to biological problems, especially in the domain of protein folding. Another equally complex and important question has received relatively little attention by the machine learning community, namely the one of prediction of complex traits from genetics. Tackling this problem requires in-depth knowledge of the related genetics literature and awareness of various subtleties associated with genetic data. In this guide, we provide an overview for the machine learning community on current state of the art models and associated subtleties which need to be taken into consideration when developing new models for phenotype prediction. We use height as an example of a continuous-valued phenotype and provide an introduction to benchmark datasets, confounders, feature selection, and common metrics.
Interpretability Aware Model Training to Improve Robustness against Out-of-Distribution Magnetic Resonance Images in Alzheimer's Disease Classification
Nov 15, 2021
Abstract:Owing to its pristine soft-tissue contrast and high resolution, structural magnetic resonance imaging (MRI) is widely applied in neurology, making it a valuable data source for image-based machine learning (ML) and deep learning applications. The physical nature of MRI acquisition and reconstruction, however, causes variations in image intensity, resolution, and signal-to-noise ratio. Since ML models are sensitive to such variations, performance on out-of-distribution data, which is inherent to the setting of a deployed healthcare ML application, typically drops below acceptable levels. We propose an interpretability aware adversarial training regime to improve robustness against out-of-distribution samples originating from different MRI hardware. The approach is applied to 1.5T and 3T MRIs obtained from the Alzheimer's Disease Neuroimaging Initiative database. We present preliminary results showing promising performance on out-of-distribution samples.
Image analysis for Alzheimer's disease prediction: Embracing pathological hallmarks for model architecture design
Nov 13, 2020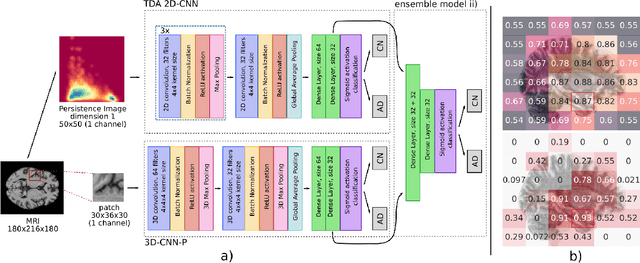
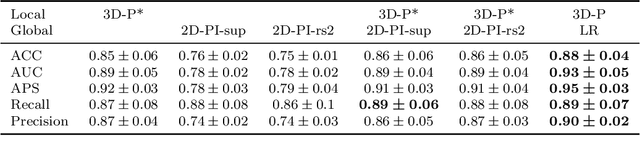
Abstract:Alzheimer's disease (AD) is associated with local (e.g. brain tissue atrophy) and global brain changes (loss of cerebral connectivity), which can be detected by high-resolution structural magnetic resonance imaging. Conventionally, these changes and their relation to AD are investigated independently. Here, we introduce a novel, highly-scalable approach that simultaneously captures $\textit{local}$ and $\textit{global}$ changes in the diseased brain. It is based on a neural network architecture that combines patch-based, high-resolution 3D-CNNs with global topological features, evaluating multi-scale brain tissue connectivity. Our local-global approach reached competitive results with an average precision score of $0.95\pm0.03$ for the classification of cognitively normal subjects and AD patients (prevalence $\approx 55\%$).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge