Boah Kim
Classification of Multi-Parametric Body MRI Series Using Deep Learning
Jun 18, 2025Abstract:Multi-parametric magnetic resonance imaging (mpMRI) exams have various series types acquired with different imaging protocols. The DICOM headers of these series often have incorrect information due to the sheer diversity of protocols and occasional technologist errors. To address this, we present a deep learning-based classification model to classify 8 different body mpMRI series types so that radiologists read the exams efficiently. Using mpMRI data from various institutions, multiple deep learning-based classifiers of ResNet, EfficientNet, and DenseNet are trained to classify 8 different MRI series, and their performance is compared. Then, the best-performing classifier is identified, and its classification capability under the setting of different training data quantities is studied. Also, the model is evaluated on the out-of-training-distribution datasets. Moreover, the model is trained using mpMRI exams obtained from different scanners in two training strategies, and its performance is tested. Experimental results show that the DenseNet-121 model achieves the highest F1-score and accuracy of 0.966 and 0.972 over the other classification models with p-value$<$0.05. The model shows greater than 0.95 accuracy when trained with over 729 studies of the training data, whose performance improves as the training data quantities grew larger. On the external data with the DLDS and CPTAC-UCEC datasets, the model yields 0.872 and 0.810 accuracy for each. These results indicate that in both the internal and external datasets, the DenseNet-121 model attains high accuracy for the task of classifying 8 body MRI series types.
Benchmarking Multi-Organ Segmentation Tools for Multi-Parametric T1-weighted Abdominal MRI
Apr 10, 2025Abstract:The segmentation of multiple organs in multi-parametric MRI studies is critical for many applications in radiology, such as correlating imaging biomarkers with disease status (e.g., cirrhosis, diabetes). Recently, three publicly available tools, such as MRSegmentator (MRSeg), TotalSegmentator MRI (TS), and TotalVibeSegmentator (VIBE), have been proposed for multi-organ segmentation in MRI. However, the performance of these tools on specific MRI sequence types has not yet been quantified. In this work, a subset of 40 volumes from the public Duke Liver Dataset was curated. The curated dataset contained 10 volumes each from the pre-contrast fat saturated T1, arterial T1w, venous T1w, and delayed T1w phases, respectively. Ten abdominal structures were manually annotated in these volumes. Next, the performance of the three public tools was benchmarked on this curated dataset. The results indicated that MRSeg obtained a Dice score of 80.7 $\pm$ 18.6 and Hausdorff Distance (HD) error of 8.9 $\pm$ 10.4 mm. It fared the best ($p < .05$) across the different sequence types in contrast to TS and VIBE.
Leveraging Multiphase CT for Quality Enhancement of Portal Venous CT: Utility for Pancreas Segmentation
Jan 23, 2025



Abstract:Multiphase CT studies are routinely obtained in clinical practice for diagnosis and management of various diseases, such as cancer. However, the CT studies can be acquired with low radiation doses, different scanners, and are frequently affected by motion and metal artifacts. Prior approaches have targeted the quality improvement of one specific CT phase (e.g., non-contrast CT). In this work, we hypothesized that leveraging multiple CT phases for the quality enhancement of one phase may prove advantageous for downstream tasks, such as segmentation. A 3D progressive fusion and non-local (PFNL) network was developed. It was trained with three degraded (low-quality) phases (non-contrast, arterial, and portal venous) to enhance the quality of the portal venous phase. Then, the effect of scan quality enhancement was evaluated using a proxy task of pancreas segmentation, which is useful for tracking pancreatic cancer. The proposed approach improved the pancreas segmentation by 3% over the corresponding low-quality CT scan. To the best of our knowledge, we are the first to harness multiphase CT for scan quality enhancement and improved pancreas segmentation.
Automated classification of multi-parametric body MRI series
May 14, 2024



Abstract:Multi-parametric MRI (mpMRI) studies are widely available in clinical practice for the diagnosis of various diseases. As the volume of mpMRI exams increases yearly, there are concomitant inaccuracies that exist within the DICOM header fields of these exams. This precludes the use of the header information for the arrangement of the different series as part of the radiologist's hanging protocol, and clinician oversight is needed for correction. In this pilot work, we propose an automated framework to classify the type of 8 different series in mpMRI studies. We used 1,363 studies acquired by three Siemens scanners to train a DenseNet-121 model with 5-fold cross-validation. Then, we evaluated the performance of the DenseNet-121 ensemble on a held-out test set of 313 mpMRI studies. Our method achieved an average precision of 96.6%, sensitivity of 96.6%, specificity of 99.6%, and F1 score of 96.6% for the MRI series classification task. To the best of our knowledge, we are the first to develop a method to classify the series type in mpMRI studies acquired at the level of the chest, abdomen, and pelvis. Our method has the capability for robust automation of hanging protocols in modern radiology practice.
MRISegmentator-Abdomen: A Fully Automated Multi-Organ and Structure Segmentation Tool for T1-weighted Abdominal MRI
May 09, 2024



Abstract:Background: Segmentation of organs and structures in abdominal MRI is useful for many clinical applications, such as disease diagnosis and radiotherapy. Current approaches have focused on delineating a limited set of abdominal structures (13 types). To date, there is no publicly available abdominal MRI dataset with voxel-level annotations of multiple organs and structures. Consequently, a segmentation tool for multi-structure segmentation is also unavailable. Methods: We curated a T1-weighted abdominal MRI dataset consisting of 195 patients who underwent imaging at National Institutes of Health (NIH) Clinical Center. The dataset comprises of axial pre-contrast T1, arterial, venous, and delayed phases for each patient, thereby amounting to a total of 780 series (69,248 2D slices). Each series contains voxel-level annotations of 62 abdominal organs and structures. A 3D nnUNet model, dubbed as MRISegmentator-Abdomen (MRISegmentator in short), was trained on this dataset, and evaluation was conducted on an internal test set and two large external datasets: AMOS22 and Duke Liver. The predicted segmentations were compared against the ground-truth using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD). Findings: MRISegmentator achieved an average DSC of 0.861$\pm$0.170 and a NSD of 0.924$\pm$0.163 in the internal test set. On the AMOS22 dataset, MRISegmentator attained an average DSC of 0.829$\pm$0.133 and a NSD of 0.908$\pm$0.067. For the Duke Liver dataset, an average DSC of 0.933$\pm$0.015 and a NSD of 0.929$\pm$0.021 was obtained. Interpretation: The proposed MRISegmentator provides automatic, accurate, and robust segmentations of 62 organs and structures in T1-weighted abdominal MRI sequences. The tool has the potential to accelerate research on various clinical topics, such as abnormality detection, radiotherapy, disease classification among others.
Automated Classification of Body MRI Sequence Type Using Convolutional Neural Networks
Feb 12, 2024Abstract:Multi-parametric MRI of the body is routinely acquired for the identification of abnormalities and diagnosis of diseases. However, a standard naming convention for the MRI protocols and associated sequences does not exist due to wide variations in imaging practice at institutions and myriad MRI scanners from various manufacturers being used for imaging. The intensity distributions of MRI sequences differ widely as a result, and there also exists information conflicts related to the sequence type in the DICOM headers. At present, clinician oversight is necessary to ensure that the correct sequence is being read and used for diagnosis. This poses a challenge when specific series need to be considered for building a cohort for a large clinical study or for developing AI algorithms. In order to reduce clinician oversight and ensure the validity of the DICOM headers, we propose an automated method to classify the 3D MRI sequence acquired at the levels of the chest, abdomen, and pelvis. In our pilot work, our 3D DenseNet-121 model achieved an F1 score of 99.5% at differentiating 5 common MRI sequences obtained by three Siemens scanners (Aera, Verio, Biograph mMR). To the best of our knowledge, we are the first to develop an automated method for the 3D classification of MRI sequences in the chest, abdomen, and pelvis, and our work has outperformed the previous state-of-the-art MRI series classifiers.
Semantic Image Synthesis for Abdominal CT
Dec 11, 2023



Abstract:As a new emerging and promising type of generative models, diffusion models have proven to outperform Generative Adversarial Networks (GANs) in multiple tasks, including image synthesis. In this work, we explore semantic image synthesis for abdominal CT using conditional diffusion models, which can be used for downstream applications such as data augmentation. We systematically evaluated the performance of three diffusion models, as well as to other state-of-the-art GAN-based approaches, and studied the different conditioning scenarios for the semantic mask. Experimental results demonstrated that diffusion models were able to synthesize abdominal CT images with better quality. Additionally, encoding the mask and the input separately is more effective than na\"ive concatenating.
C-DARL: Contrastive diffusion adversarial representation learning for label-free blood vessel segmentation
Jul 31, 2023Abstract:Blood vessel segmentation in medical imaging is one of the essential steps for vascular disease diagnosis and interventional planning in a broad spectrum of clinical scenarios in image-based medicine and interventional medicine. Unfortunately, manual annotation of the vessel masks is challenging and resource-intensive due to subtle branches and complex structures. To overcome this issue, this paper presents a self-supervised vessel segmentation method, dubbed the contrastive diffusion adversarial representation learning (C-DARL) model. Our model is composed of a diffusion module and a generation module that learns the distribution of multi-domain blood vessel data by generating synthetic vessel images from diffusion latent. Moreover, we employ contrastive learning through a mask-based contrastive loss so that the model can learn more realistic vessel representations. To validate the efficacy, C-DARL is trained using various vessel datasets, including coronary angiograms, abdominal digital subtraction angiograms, and retinal imaging. Experimental results confirm that our model achieves performance improvement over baseline methods with noise robustness, suggesting the effectiveness of C-DARL for vessel segmentation.
Diffusion Adversarial Representation Learning for Self-supervised Vessel Segmentation
Sep 29, 2022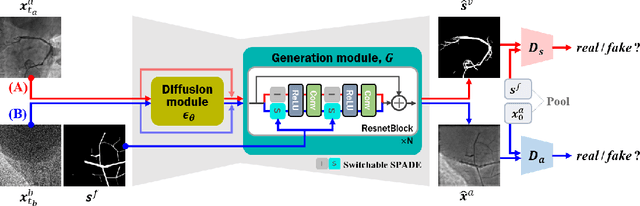
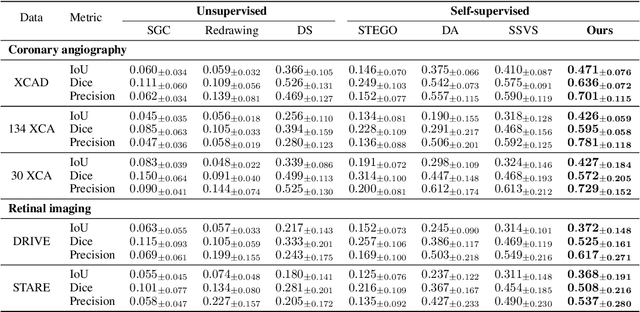
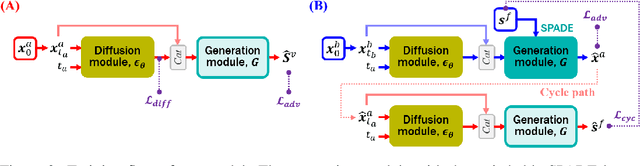
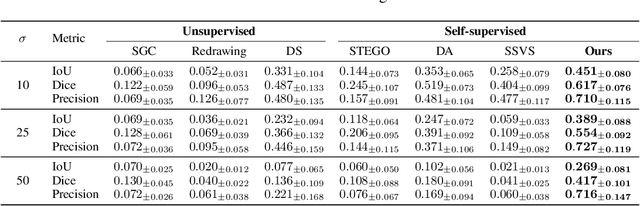
Abstract:Vessel segmentation in medical images is one of the important tasks in the diagnosis of vascular diseases and therapy planning. Although learning-based segmentation approaches have been extensively studied, a large amount of ground-truth labels are required in supervised methods and confusing background structures make neural networks hard to segment vessels in an unsupervised manner. To address this, here we introduce a novel diffusion adversarial representation learning (DARL) model that leverages a denoising diffusion probabilistic model with adversarial learning, and apply it for vessel segmentation. In particular, for self-supervised vessel segmentation, DARL learns background image distribution using a diffusion module, which lets a generation module effectively provide vessel representations. Also, by adversarial learning based on the proposed switchable spatially-adaptive denormalization, our model estimates synthetic fake vessel images as well as vessel segmentation masks, which further makes the model capture vessel-relevant semantic information. Once the proposed model is trained, the model generates segmentation masks by one step and can be applied to general vascular structure segmentation of coronary angiography and retinal images. Experimental results on various datasets show that our method significantly outperforms existing unsupervised and self-supervised methods in vessel segmentation.
Diffusion Deformable Model for 4D Temporal Medical Image Generation
Jun 27, 2022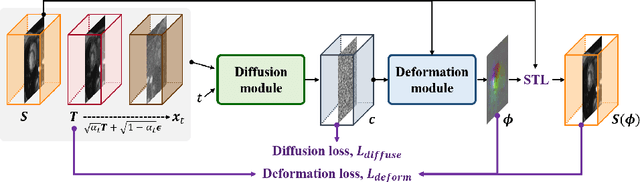
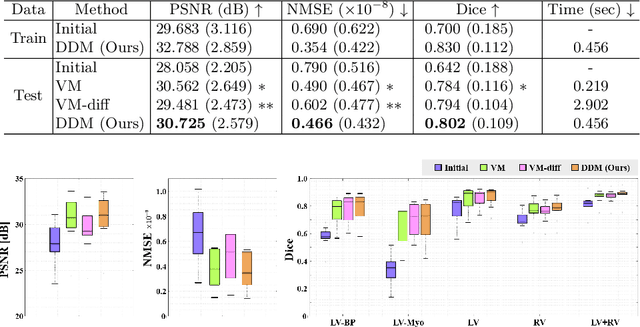
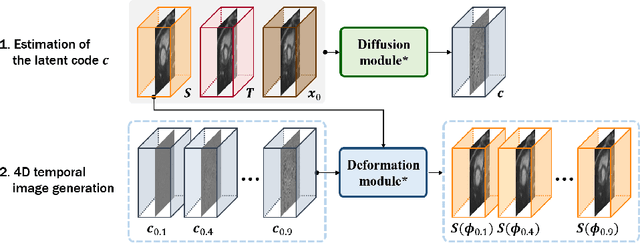
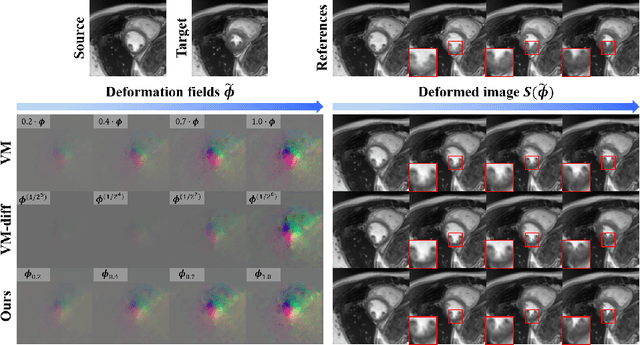
Abstract:Temporal volume images with 3D+t (4D) information are often used in medical imaging to statistically analyze temporal dynamics or capture disease progression. Although deep-learning-based generative models for natural images have been extensively studied, approaches for temporal medical image generation such as 4D cardiac volume data are limited. In this work, we present a novel deep learning model that generates intermediate temporal volumes between source and target volumes. Specifically, we propose a diffusion deformable model (DDM) by adapting the denoising diffusion probabilistic model that has recently been widely investigated for realistic image generation. Our proposed DDM is composed of the diffusion and the deformation modules so that DDM can learn spatial deformation information between the source and target volumes and provide a latent code for generating intermediate frames along a geodesic path. Once our model is trained, the latent code estimated from the diffusion module is simply interpolated and fed into the deformation module, which enables DDM to generate temporal frames along the continuous trajectory while preserving the topology of the source image. We demonstrate the proposed method with the 4D cardiac MR image generation between the diastolic and systolic phases for each subject. Compared to the existing deformation methods, our DDM achieves high performance on temporal volume generation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge