Tejas Sudharshan Mathai
Utility of Pancreas Surface Lobularity as a CT Biomarker for Opportunistic Screening of Type 2 Diabetes
Nov 13, 2025


Abstract:Type 2 Diabetes Mellitus (T2DM) is a chronic metabolic disease that affects millions of people worldwide. Early detection is crucial as it can alter pancreas function through morphological changes and increased deposition of ectopic fat, eventually leading to organ damage. While studies have shown an association between T2DM and pancreas volume and fat content, the role of increased pancreatic surface lobularity (PSL) in patients with T2DM has not been fully investigated. In this pilot work, we propose a fully automated approach to delineate the pancreas and other abdominal structures, derive CT imaging biomarkers, and opportunistically screen for T2DM. Four deep learning-based models were used to segment the pancreas in an internal dataset of 584 patients (297 males, 437 non-diabetic, age: 45$\pm$15 years). PSL was automatically detected and it was higher for diabetic patients (p=0.01) at 4.26 $\pm$ 8.32 compared to 3.19 $\pm$ 3.62 for non-diabetic patients. The PancAP model achieved the highest Dice score of 0.79 $\pm$ 0.17 and lowest ASSD error of 1.94 $\pm$ 2.63 mm (p$<$0.05). For predicting T2DM, a multivariate model trained with CT biomarkers attained 0.90 AUC, 66.7\% sensitivity, and 91.9\% specificity. Our results suggest that PSL is useful for T2DM screening and could potentially help predict the early onset of T2DM.
Text Embedded Swin-UMamba for DeepLesion Segmentation
Aug 08, 2025Abstract:Segmentation of lesions on CT enables automatic measurement for clinical assessment of chronic diseases (e.g., lymphoma). Integrating large language models (LLMs) into the lesion segmentation workflow offers the potential to combine imaging features with descriptions of lesion characteristics from the radiology reports. In this study, we investigate the feasibility of integrating text into the Swin-UMamba architecture for the task of lesion segmentation. The publicly available ULS23 DeepLesion dataset was used along with short-form descriptions of the findings from the reports. On the test dataset, a high Dice Score of 82% and low Hausdorff distance of 6.58 (pixels) was obtained for lesion segmentation. The proposed Text-Swin-UMamba model outperformed prior approaches: 37% improvement over the LLM-driven LanGuideMedSeg model (p < 0.001),and surpassed the purely image-based xLSTM-UNet and nnUNet models by 1.74% and 0.22%, respectively. The dataset and code can be accessed at https://github.com/ruida/LLM-Swin-UMamba
Classification of Multi-Parametric Body MRI Series Using Deep Learning
Jun 18, 2025Abstract:Multi-parametric magnetic resonance imaging (mpMRI) exams have various series types acquired with different imaging protocols. The DICOM headers of these series often have incorrect information due to the sheer diversity of protocols and occasional technologist errors. To address this, we present a deep learning-based classification model to classify 8 different body mpMRI series types so that radiologists read the exams efficiently. Using mpMRI data from various institutions, multiple deep learning-based classifiers of ResNet, EfficientNet, and DenseNet are trained to classify 8 different MRI series, and their performance is compared. Then, the best-performing classifier is identified, and its classification capability under the setting of different training data quantities is studied. Also, the model is evaluated on the out-of-training-distribution datasets. Moreover, the model is trained using mpMRI exams obtained from different scanners in two training strategies, and its performance is tested. Experimental results show that the DenseNet-121 model achieves the highest F1-score and accuracy of 0.966 and 0.972 over the other classification models with p-value$<$0.05. The model shows greater than 0.95 accuracy when trained with over 729 studies of the training data, whose performance improves as the training data quantities grew larger. On the external data with the DLDS and CPTAC-UCEC datasets, the model yields 0.872 and 0.810 accuracy for each. These results indicate that in both the internal and external datasets, the DenseNet-121 model attains high accuracy for the task of classifying 8 body MRI series types.
Benchmarking Multi-Organ Segmentation Tools for Multi-Parametric T1-weighted Abdominal MRI
Apr 10, 2025Abstract:The segmentation of multiple organs in multi-parametric MRI studies is critical for many applications in radiology, such as correlating imaging biomarkers with disease status (e.g., cirrhosis, diabetes). Recently, three publicly available tools, such as MRSegmentator (MRSeg), TotalSegmentator MRI (TS), and TotalVibeSegmentator (VIBE), have been proposed for multi-organ segmentation in MRI. However, the performance of these tools on specific MRI sequence types has not yet been quantified. In this work, a subset of 40 volumes from the public Duke Liver Dataset was curated. The curated dataset contained 10 volumes each from the pre-contrast fat saturated T1, arterial T1w, venous T1w, and delayed T1w phases, respectively. Ten abdominal structures were manually annotated in these volumes. Next, the performance of the three public tools was benchmarked on this curated dataset. The results indicated that MRSeg obtained a Dice score of 80.7 $\pm$ 18.6 and Hausdorff Distance (HD) error of 8.9 $\pm$ 10.4 mm. It fared the best ($p < .05$) across the different sequence types in contrast to TS and VIBE.
Longitudinal Assessment of Lung Lesion Burden in CT
Apr 09, 2025



Abstract:In the U.S., lung cancer is the second major cause of death. Early detection of suspicious lung nodules is crucial for patient treatment planning, management, and improving outcomes. Many approaches for lung nodule segmentation and volumetric analysis have been proposed, but few have looked at longitudinal changes in total lung tumor burden. In this work, we trained two 3D models (nnUNet) with and without anatomical priors to automatically segment lung lesions and quantified total lesion burden for each patient. The 3D model without priors significantly outperformed ($p < .001$) the model trained with anatomy priors. For detecting clinically significant lesions $>$ 1cm, a precision of 71.3\%, sensitivity of 68.4\%, and F1-score of 69.8\% was achieved. For segmentation, a Dice score of 77.1 $\pm$ 20.3 and Hausdorff distance error of 11.7 $\pm$ 24.1 mm was obtained. The median lesion burden was 6.4 cc (IQR: 2.1, 18.1) and the median volume difference between manual and automated measurements was 0.02 cc (IQR: -2.8, 1.2). Agreements were also evaluated with linear regression and Bland-Altman plots. The proposed approach can produce a personalized evaluation of the total tumor burden for a patient and facilitate interval change tracking over time.
Leveraging Anatomical Priors for Automated Pancreas Segmentation on Abdominal CT
Apr 09, 2025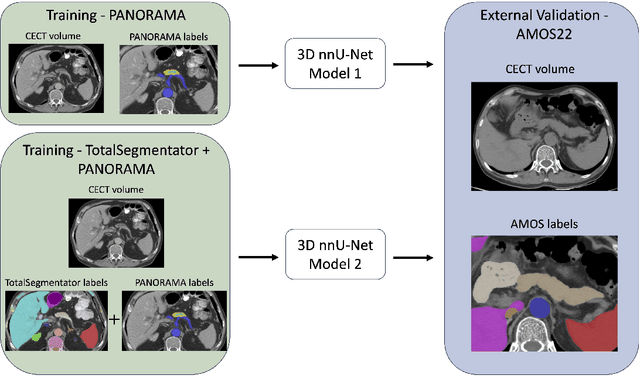


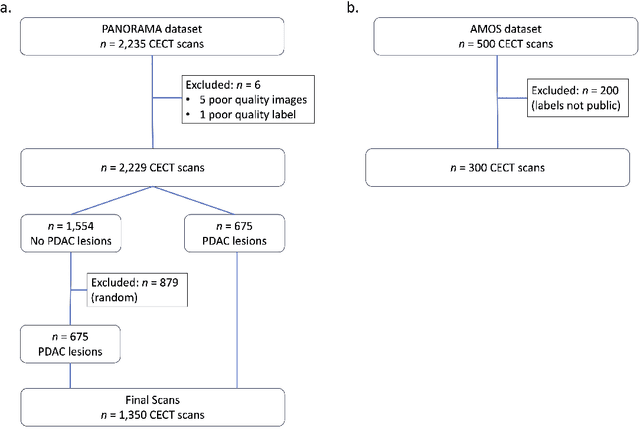
Abstract:An accurate segmentation of the pancreas on CT is crucial to identify pancreatic pathologies and extract imaging-based biomarkers. However, prior research on pancreas segmentation has primarily focused on modifying the segmentation model architecture or utilizing pre- and post-processing techniques. In this article, we investigate the utility of anatomical priors to enhance the segmentation performance of the pancreas. Two 3D full-resolution nnU-Net models were trained, one with 8 refined labels from the public PANORAMA dataset, and another that combined them with labels derived from the public TotalSegmentator (TS) tool. The addition of anatomical priors resulted in a 6\% increase in Dice score ($p < .001$) and a 36.5 mm decrease in Hausdorff distance for pancreas segmentation ($p < .001$). Moreover, the pancreas was always detected when anatomy priors were used, whereas there were 8 instances of failed detections without their use. The use of anatomy priors shows promise for pancreas segmentation and subsequent derivation of imaging biomarkers.
Class Imbalance Correction for Improved Universal Lesion Detection and Tagging in CT
Apr 08, 2025Abstract:Radiologists routinely detect and size lesions in CT to stage cancer and assess tumor burden. To potentially aid their efforts, multiple lesion detection algorithms have been developed with a large public dataset called DeepLesion (32,735 lesions, 32,120 CT slices, 10,594 studies, 4,427 patients, 8 body part labels). However, this dataset contains missing measurements and lesion tags, and exhibits a severe imbalance in the number of lesions per label category. In this work, we utilize a limited subset of DeepLesion (6\%, 1331 lesions, 1309 slices) containing lesion annotations and body part label tags to train a VFNet model to detect lesions and tag them. We address the class imbalance by conducting three experiments: 1) Balancing data by the body part labels, 2) Balancing data by the number of lesions per patient, and 3) Balancing data by the lesion size. In contrast to a randomly sampled (unbalanced) data subset, our results indicated that balancing the body part labels always increased sensitivity for lesions >= 1cm for classes with low data quantities (Bone: 80\% vs. 46\%, Kidney: 77\% vs. 61\%, Soft Tissue: 70\% vs. 60\%, Pelvis: 83\% vs. 76\%). Similar trends were seen for three other models tested (FasterRCNN, RetinaNet, FoveaBox). Balancing data by lesion size also helped the VFNet model improve recalls for all classes in contrast to an unbalanced dataset. We also provide a structured reporting guideline for a ``Lesions'' subsection to be entered into the ``Findings'' section of a radiology report. To our knowledge, we are the first to report the class imbalance in DeepLesion, and have taken data-driven steps to address it in the context of joint lesion detection and tagging.
3D Universal Lesion Detection and Tagging in CT with Self-Training
Apr 07, 2025Abstract:Radiologists routinely perform the tedious task of lesion localization, classification, and size measurement in computed tomography (CT) studies. Universal lesion detection and tagging (ULDT) can simultaneously help alleviate the cumbersome nature of lesion measurement and enable tumor burden assessment. Previous ULDT approaches utilize the publicly available DeepLesion dataset, however it does not provide the full volumetric (3D) extent of lesions and also displays a severe class imbalance. In this work, we propose a self-training pipeline to detect 3D lesions and tag them according to the body part they occur in. We used a significantly limited 30\% subset of DeepLesion to train a VFNet model for 2D lesion detection and tagging. Next, the 2D lesion context was expanded into 3D, and the mined 3D lesion proposals were integrated back into the baseline training data in order to retrain the model over multiple rounds. Through the self-training procedure, our VFNet model learned from its own predictions, detected lesions in 3D, and tagged them. Our results indicated that our VFNet model achieved an average sensitivity of 46.9\% at [0.125:8] false positives (FP) with a limited 30\% data subset in comparison to the 46.8\% of an existing approach that used the entire DeepLesion dataset. To our knowledge, we are the first to jointly detect lesions in 3D and tag them according to the body part label.
Universal Lymph Node Detection in Multiparametric MRI with Selective Augmentation
Apr 07, 2025Abstract:Robust localization of lymph nodes (LNs) in multiparametric MRI (mpMRI) is critical for the assessment of lymphadenopathy. Radiologists routinely measure the size of LN to distinguish benign from malignant nodes, which would require subsequent cancer staging. Sizing is a cumbersome task compounded by the diverse appearances of LNs in mpMRI, which renders their measurement difficult. Furthermore, smaller and potentially metastatic LNs could be missed during a busy clinical day. To alleviate these imaging and workflow problems, we propose a pipeline to universally detect both benign and metastatic nodes in the body for their ensuing measurement. The recently proposed VFNet neural network was employed to identify LN in T2 fat suppressed and diffusion weighted imaging (DWI) sequences acquired by various scanners with a variety of exam protocols. We also use a selective augmentation technique known as Intra-Label LISA (ILL) to diversify the input data samples the model sees during training, such that it improves its robustness during the evaluation phase. We achieved a sensitivity of $\sim$83\% with ILL vs. $\sim$80\% without ILL at 4 FP/vol. Compared with current LN detection approaches evaluated on mpMRI, we show a sensitivity improvement of $\sim$9\% at 4 FP/vol.
Correcting Class Imbalances with Self-Training for Improved Universal Lesion Detection and Tagging
Apr 07, 2025Abstract:Universal lesion detection and tagging (ULDT) in CT studies is critical for tumor burden assessment and tracking the progression of lesion status (growth/shrinkage) over time. However, a lack of fully annotated data hinders the development of effective ULDT approaches. Prior work used the DeepLesion dataset (4,427 patients, 10,594 studies, 32,120 CT slices, 32,735 lesions, 8 body part labels) for algorithmic development, but this dataset is not completely annotated and contains class imbalances. To address these issues, in this work, we developed a self-training pipeline for ULDT. A VFNet model was trained on a limited 11.5\% subset of DeepLesion (bounding boxes + tags) to detect and classify lesions in CT studies. Then, it identified and incorporated novel lesion candidates from a larger unseen data subset into its training set, and self-trained itself over multiple rounds. Multiple self-training experiments were conducted with different threshold policies to select predicted lesions with higher quality and cover the class imbalances. We discovered that direct self-training improved the sensitivities of over-represented lesion classes at the expense of under-represented classes. However, upsampling the lesions mined during self-training along with a variable threshold policy yielded a 6.5\% increase in sensitivity at 4 FP in contrast to self-training without class balancing (72\% vs 78.5\%) and a 11.7\% increase compared to the same self-training policy without upsampling (66.8\% vs 78.5\%). Furthermore, we show that our results either improved or maintained the sensitivity at 4FP for all 8 lesion classes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge