Andrea Mastropietro
VitaGraph: Building a Knowledge Graph for Biologically Relevant Learning Tasks
May 16, 2025Abstract:The intrinsic complexity of human biology presents ongoing challenges to scientific understanding. Researchers collaborate across disciplines to expand our knowledge of the biological interactions that define human life. AI methodologies have emerged as powerful tools across scientific domains, particularly in computational biology, where graph data structures effectively model biological entities such as protein-protein interaction (PPI) networks and gene functional networks. Those networks are used as datasets for paramount network medicine tasks, such as gene-disease association prediction, drug repurposing, and polypharmacy side effect studies. Reliable predictions from machine learning models require high-quality foundational data. In this work, we present a comprehensive multi-purpose biological knowledge graph constructed by integrating and refining multiple publicly available datasets. Building upon the Drug Repurposing Knowledge Graph (DRKG), we define a pipeline tasked with a) cleaning inconsistencies and redundancies present in DRKG, b) coalescing information from the main available public data sources, and c) enriching the graph nodes with expressive feature vectors such as molecular fingerprints and gene ontologies. Biologically and chemically relevant features improve the capacity of machine learning models to generate accurate and well-structured embedding spaces. The resulting resource represents a coherent and reliable biological knowledge graph that serves as a state-of-the-art platform to advance research in computational biology and precision medicine. Moreover, it offers the opportunity to benchmark graph-based machine learning and network medicine models on relevant tasks. We demonstrate the effectiveness of the proposed dataset by benchmarking it against the task of drug repurposing, PPI prediction, and side-effect prediction, modeled as link prediction problems.
Kolmogorov-Arnold Graph Neural Networks
Jun 26, 2024Abstract:Graph neural networks (GNNs) excel in learning from network-like data but often lack interpretability, making their application challenging in domains requiring transparent decision-making. We propose the Graph Kolmogorov-Arnold Network (GKAN), a novel GNN model leveraging spline-based activation functions on edges to enhance both accuracy and interpretability. Our experiments on five benchmark datasets demonstrate that GKAN outperforms state-of-the-art GNN models in node classification, link prediction, and graph classification tasks. In addition to the improved accuracy, GKAN's design inherently provides clear insights into the model's decision-making process, eliminating the need for post-hoc explainability techniques. This paper discusses the methodology, performance, and interpretability of GKAN, highlighting its potential for applications in domains where interpretability is crucial.
AI-based Data Preparation and Data Analytics in Healthcare: The Case of Diabetes
Jun 13, 2022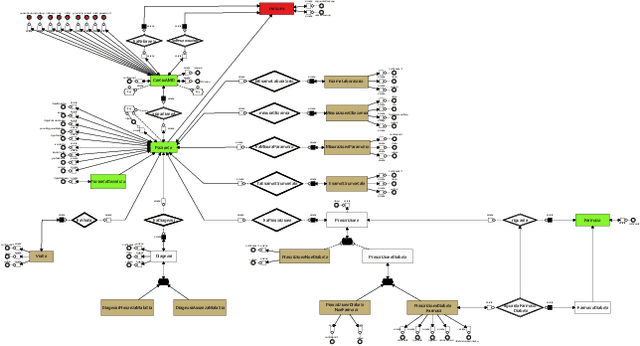
Abstract:The Associazione Medici Diabetologi (AMD) collects and manages one of the largest worldwide-available collections of diabetic patient records, also known as the AMD database. This paper presents the initial results of an ongoing project whose focus is the application of Artificial Intelligence and Machine Learning techniques for conceptualizing, cleaning, and analyzing such an important and valuable dataset, with the goal of providing predictive insights to better support diabetologists in their diagnostic and therapeutic choices.
Adaptive Positive-Unlabelled Learning via Markov Diffusion
Aug 13, 2021
Abstract:Positive-Unlabelled (PU) learning is the machine learning setting in which only a set of positive instances are labelled, while the rest of the data set is unlabelled. The unlabelled instances may be either unspecified positive samples or true negative samples. Over the years, many solutions have been proposed to deal with PU learning. Some techniques consider the unlabelled samples as negative ones, reducing the problem to a binary classification with a noisy negative set, while others aim to detect sets of possible negative examples to later apply a supervised machine learning strategy (two-step techniques). The approach proposed in this work falls in the latter category and works in a semi-supervised fashion: motivated and inspired by previous works, a Markov diffusion process with restart is used to assign pseudo-labels to unlabelled instances. Afterward, a machine learning model, exploiting the newly assigned classes, is trained. The principal aim of the algorithm is to identify a set of instances which are likely to contain positive instances that were originally unlabelled.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge