Andrea Gondova
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


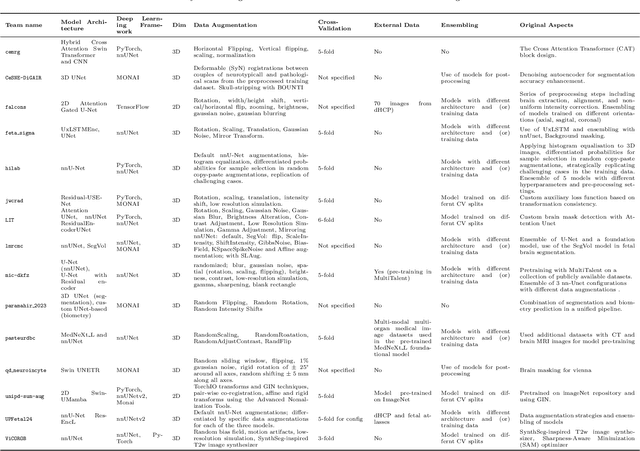
Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Towards Fully Automated Segmentation of Rat Cardiac MRI by Leveraging Deep Learning Frameworks
Sep 09, 2021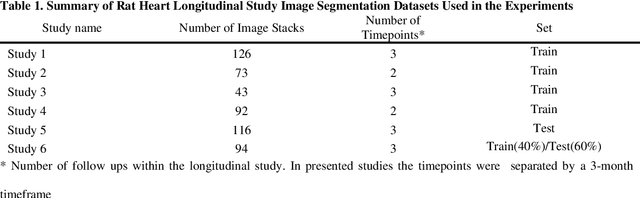
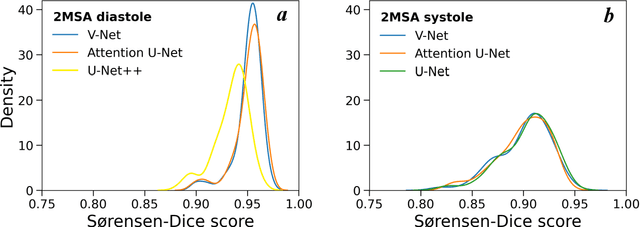

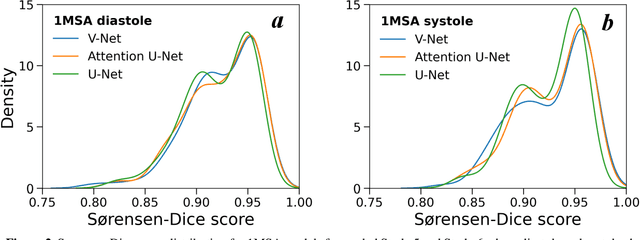
Abstract:Automated segmentation of human cardiac magnetic resonance datasets has been steadily improving during recent years. However, these methods are not directly applicable in preclinical context due to limited datasets and lower image resolution. Successful application of deep architectures for rat cardiac segmentation, although of critical importance for preclinical evaluation of cardiac function, has to our knowledge not yet been reported. We developed segmentation models that expand on the standard U-Net architecture and evaluated separate models for systole and diastole phases, 2MSA, and one model for all timepoints, 1MSA. Furthermore, we calibrated model outputs using a Gaussian Process (GP)-based prior to improve phase selection. Resulting models approach human performance in terms of left ventricular segmentation quality and ejection fraction (EF) estimation in both 1MSA and 2MSA settings (S{\o}rensen-Dice score 0.91 +/- 0.072 and 0.93 +/- 0.032, respectively). 2MSA achieved a mean absolute difference between estimated and reference EF of 3.5 +/- 2.5 %, while 1MSA resulted in 4.1 +/- 3.0 %. Applying Gaussian Processes to 1MSA allows to automate the selection of systole and diastole phases. Combined with a novel cardiac phase selection strategy, our work presents an important first step towards a fully automated segmentation pipeline in the context of rat cardiac analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge