Aditya Pola
LogicCBMs: Logic-Enhanced Concept-Based Learning
Dec 08, 2025



Abstract:Concept Bottleneck Models (CBMs) provide a basis for semantic abstractions within a neural network architecture. Such models have primarily been seen through the lens of interpretability so far, wherein they offer transparency by inferring predictions as a linear combination of semantic concepts. However, a linear combination is inherently limiting. So we propose the enhancement of concept-based learning models through propositional logic. We introduce a logic module that is carefully designed to connect the learned concepts from CBMs through differentiable logic operations, such that our proposed LogicCBM can go beyond simple weighted combinations of concepts to leverage various logical operations to yield the final predictions, while maintaining end-to-end learnability. Composing concepts using a set of logic operators enables the model to capture inter-concept relations, while simultaneously improving the expressivity of the model in terms of logic operations. Our empirical studies on well-known benchmarks and synthetic datasets demonstrate that these models have better accuracy, perform effective interventions and are highly interpretable.
Where does an LLM begin computing an instruction?
Nov 19, 2025Abstract:Following an instruction involves distinct sub-processes, such as reading content, reading the instruction, executing it, and producing an answer. We ask where, along the layer stack, instruction following begins, the point where reading gives way to doing. We introduce three simple datasets (Key-Value, Quote Attribution, Letter Selection) and two hop compositions of these tasks. Using activation patching on minimal-contrast prompt pairs, we measure a layer-wise flip rate that indicates when substituting selected residual activations changes the predicted answer. Across models in the Llama family, we observe an inflection point, which we term onset, where interventions that change predictions before this point become largely ineffective afterward. Multi-hop compositions show a similar onset location. These results provide a simple, replicable way to locate where instruction following begins and to compare this location across tasks and model sizes.
LogGENE: A smooth alternative to check loss for Deep Healthcare Inference Tasks
Jun 19, 2022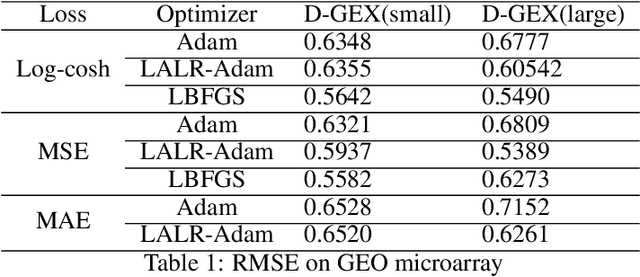
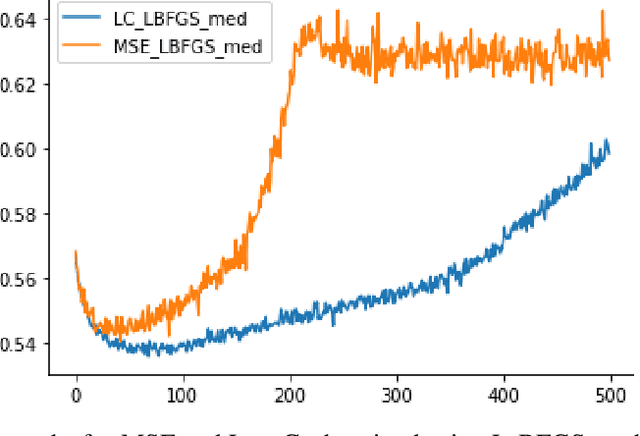
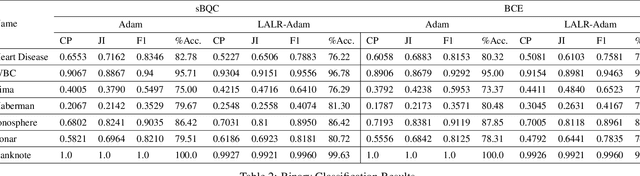
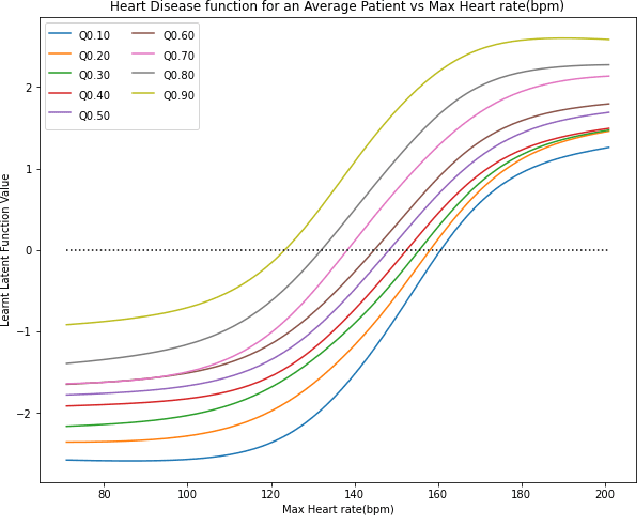
Abstract:High-throughput Genomics is ushering a new era in personalized health care, and targeted drug design and delivery. Mining these large datasets, and obtaining calibrated predictions is of immediate relevance and utility. In our work, we develop methods for Gene Expression Inference based on Deep neural networks. However, unlike typical Deep learning methods, our inferential technique, while achieving state-of-the-art performance in terms of accuracy, can also provide explanations, and report uncertainty estimates. We adopt the Quantile Regression framework to predict full conditional quantiles for a given set of house keeping gene expressions. Conditional quantiles, in addition to being useful in providing rich interpretations of the predictions, are also robust to measurement noise. However, check loss, used in quantile regression to drive the estimation process is not differentiable. We propose log-cosh as a smooth-alternative to the check loss. We apply our methods on GEO microarray dataset. We also extend the method to binary classification setting. Furthermore, we investigate other consequences of the smoothness of the loss in faster convergence.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge