Óscar Escudero-Arnanz
Early Detection of Multidrug Resistance Using Multivariate Time Series Analysis and Interpretable Patient-Similarity Representations
Apr 24, 2025Abstract:Background and Objectives: Multidrug Resistance (MDR) is a critical global health issue, causing increased hospital stays, healthcare costs, and mortality. This study proposes an interpretable Machine Learning (ML) framework for MDR prediction, aiming for both accurate inference and enhanced explainability. Methods: Patients are modeled as Multivariate Time Series (MTS), capturing clinical progression and patient-to-patient interactions. Similarity among patients is quantified using MTS-based methods: descriptive statistics, Dynamic Time Warping, and Time Cluster Kernel. These similarity measures serve as inputs for MDR classification via Logistic Regression, Random Forest, and Support Vector Machines, with dimensionality reduction and kernel transformations improving model performance. For explainability, patient similarity networks are constructed from these metrics. Spectral clustering and t-SNE are applied to identify MDR-related subgroups and visualize high-risk clusters, enabling insight into clinically relevant patterns. Results: The framework was validated on ICU Electronic Health Records from the University Hospital of Fuenlabrada, achieving an AUC of 81%. It outperforms baseline ML and deep learning models by leveraging graph-based patient similarity. The approach identifies key risk factors -- prolonged antibiotic use, invasive procedures, co-infections, and extended ICU stays -- and reveals clinically meaningful clusters. Code and results are available at \https://github.com/oscarescuderoarnanz/DM4MTS. Conclusions: Patient similarity representations combined with graph-based analysis provide accurate MDR prediction and interpretable insights. This method supports early detection, risk factor identification, and patient stratification, highlighting the potential of explainable ML in critical care.
Explainable Spatio-Temporal GCNNs for Irregular Multivariate Time Series: Architecture and Application to ICU Patient Data
Nov 01, 2024



Abstract:In this paper, we present XST-GCNN (eXplainable Spatio-Temporal Graph Convolutional Neural Network), a novel architecture for processing heterogeneous and irregular Multivariate Time Series (MTS) data. Our approach captures temporal and feature dependencies within a unified spatio-temporal pipeline by leveraging a GCNN that uses a spatio-temporal graph aimed at optimizing predictive accuracy and interoperability. For graph estimation, we introduce techniques, including one based on the (heterogeneous) Gower distance. Once estimated, we propose two methods for graph construction: one based on the Cartesian product, treating temporal instants homogeneously, and another spatio-temporal approach with distinct graphs per time step. We also propose two GCNN architectures: a standard GCNN with a normalized adjacency matrix and a higher-order polynomial GCNN. In addition to accuracy, we emphasize explainability by designing an inherently interpretable model and performing a thorough interpretability analysis, identifying key feature-time combinations that drive predictions. We evaluate XST-GCNN using real-world Electronic Health Record data from University Hospital of Fuenlabrada to predict Multidrug Resistance (MDR) in ICU patients, a critical healthcare challenge linked to high mortality and complex treatments. Our architecture outperforms traditional models, achieving a mean ROC-AUC score of 81.03 +- 2.43. Furthermore, the interpretability analysis provides actionable insights into clinical factors driving MDR predictions, enhancing model transparency. This work sets a benchmark for tackling complex inference tasks with heterogeneous MTS, offering a versatile, interpretable solution for real-world applications.
On the Use of Time Series Kernel and Dimensionality Reduction to Identify the Acquisition of Antimicrobial Multidrug Resistance in the Intensive Care Unit
Jul 07, 2021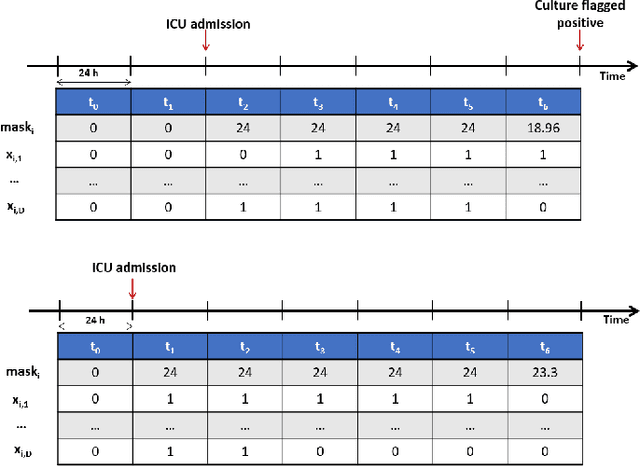
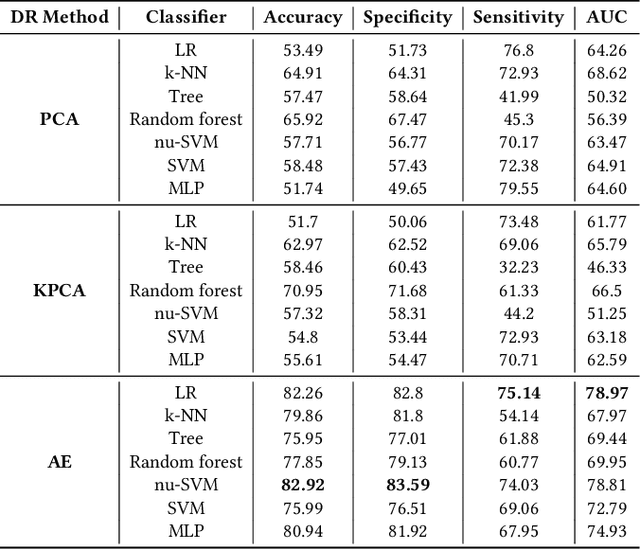
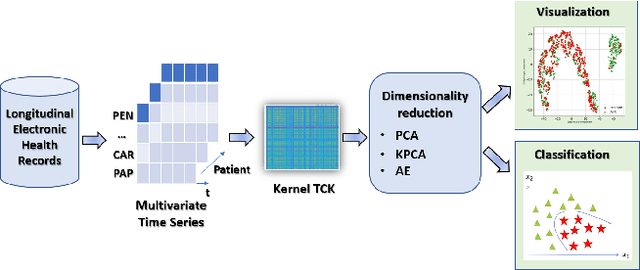
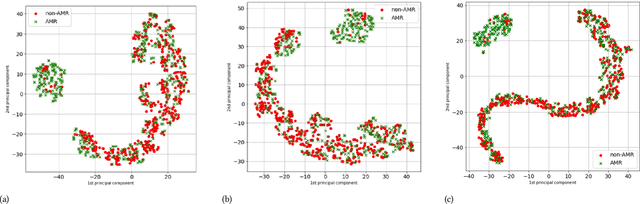
Abstract:The acquisition of Antimicrobial Multidrug Resistance (AMR) in patients admitted to the Intensive Care Units (ICU) is a major global concern. This study analyses data in the form of multivariate time series (MTS) from 3476 patients recorded at the ICU of University Hospital of Fuenlabrada (Madrid) from 2004 to 2020. 18\% of the patients acquired AMR during their stay in the ICU. The goal of this paper is an early prediction of the development of AMR. Towards that end, we leverage the time-series cluster kernel (TCK) to learn similarities between MTS. To evaluate the effectiveness of TCK as a kernel, we applied several dimensionality reduction techniques for visualization and classification tasks. The experimental results show that TCK allows identifying a group of patients that acquire the AMR during the first 48 hours of their ICU stay, and it also provides good classification capabilities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge