cancer detection
Cancer detection using Artificial Intelligence (AI) involves leveraging advanced machine learning algorithms and techniques to identify and diagnose cancer from various medical data sources. The goal is to enhance early detection, improve diagnostic accuracy, and potentially reduce the need for invasive procedures.
Papers and Code
MeisenMeister: A Simple Two Stage Pipeline for Breast Cancer Classification on MRI
Oct 31, 2025The ODELIA Breast MRI Challenge 2025 addresses a critical issue in breast cancer screening: improving early detection through more efficient and accurate interpretation of breast MRI scans. Even though methods for general-purpose whole-body lesion segmentation as well as multi-time-point analysis exist, breast cancer detection remains highly challenging, largely due to the limited availability of high-quality segmentation labels. Therefore, developing robust classification-based approaches is crucial for the future of early breast cancer detection, particularly in applications such as large-scale screening. In this write-up, we provide a comprehensive overview of our approach to the challenge. We begin by detailing the underlying concept and foundational assumptions that guided our work. We then describe the iterative development process, highlighting the key stages of experimentation, evaluation, and refinement that shaped the evolution of our solution. Finally, we present the reasoning and evidence that informed the design choices behind our final submission, with a focus on performance, robustness, and clinical relevance. We release our full implementation publicly at https://github.com/MIC-DKFZ/MeisenMeister
Adapted Foundation Models for Breast MRI Triaging in Contrast-Enhanced and Non-Contrast Enhanced Protocols
Nov 08, 2025Background: Magnetic resonance imaging (MRI) has high sensitivity for breast cancer detection, but interpretation is time-consuming. Artificial intelligence may aid in pre-screening. Purpose: To evaluate the DINOv2-based Medical Slice Transformer (MST) for ruling out significant findings (Breast Imaging Reporting and Data System [BI-RADS] >=4) in contrast-enhanced and non-contrast-enhanced abbreviated breast MRI. Materials and Methods: This institutional review board approved retrospective study included 1,847 single-breast MRI examinations (377 BI-RADS >=4) from an in-house dataset and 924 from an external validation dataset (Duke). Four abbreviated protocols were tested: T1-weighted early subtraction (T1sub), diffusion-weighted imaging with b=1500 s/mm2 (DWI1500), DWI1500+T2-weighted (T2w), and T1sub+T2w. Performance was assessed at 90%, 95%, and 97.5% sensitivity using five-fold cross-validation and area under the receiver operating characteristic curve (AUC) analysis. AUC differences were compared with the DeLong test. False negatives were characterized, and attention maps of true positives were rated in the external dataset. Results: A total of 1,448 female patients (mean age, 49 +/- 12 years) were included. T1sub+T2w achieved an AUC of 0.77 +/- 0.04; DWI1500+T2w, 0.74 +/- 0.04 (p=0.15). At 97.5% sensitivity, T1sub+T2w had the highest specificity (19% +/- 7%), followed by DWI1500+T2w (17% +/- 11%). Missed lesions had a mean diameter <10 mm at 95% and 97.5% thresholds for both T1sub and DWI1500, predominantly non-mass enhancements. External validation yielded an AUC of 0.77, with 88% of attention maps rated good or moderate. Conclusion: At 97.5% sensitivity, the MST framework correctly triaged cases without BI-RADS >=4, achieving 19% specificity for contrast-enhanced and 17% for non-contrast-enhanced MRI. Further research is warranted before clinical implementation.
H-CNN-ViT: A Hierarchical Gated Attention Multi-Branch Model for Bladder Cancer Recurrence Prediction
Nov 19, 2025Bladder cancer is one of the most prevalent malignancies worldwide, with a recurrence rate of up to 78%, necessitating accurate post-operative monitoring for effective patient management. Multi-sequence contrast-enhanced MRI is commonly used for recurrence detection; however, interpreting these scans remains challenging, even for experienced radiologists, due to post-surgical alterations such as scarring, swelling, and tissue remodeling. AI-assisted diagnostic tools have shown promise in improving bladder cancer recurrence prediction, yet progress in this field is hindered by the lack of dedicated multi-sequence MRI datasets for recurrence assessment study. In this work, we first introduce a curated multi-sequence, multi-modal MRI dataset specifically designed for bladder cancer recurrence prediction, establishing a valuable benchmark for future research. We then propose H-CNN-ViT, a new Hierarchical Gated Attention Multi-Branch model that enables selective weighting of features from the global (ViT) and local (CNN) paths based on contextual demands, achieving a balanced and targeted feature fusion. Our multi-branch architecture processes each modality independently, ensuring that the unique properties of each imaging channel are optimally captured and integrated. Evaluated on our dataset, H-CNN-ViT achieves an AUC of 78.6%, surpassing state-of-the-art models. Our model is publicly available at https://github.com/XLIAaron/H-CNN-ViT.
MedFedPure: A Medical Federated Framework with MAE-based Detection and Diffusion Purification for Inference-Time Attacks
Nov 07, 2025Artificial intelligence (AI) has shown great potential in medical imaging, particularly for brain tumor detection using Magnetic Resonance Imaging (MRI). However, the models remain vulnerable at inference time when they are trained collaboratively through Federated Learning (FL), an approach adopted to protect patient privacy. Adversarial attacks can subtly alter medical scans in ways invisible to the human eye yet powerful enough to mislead AI models, potentially causing serious misdiagnoses. Existing defenses often assume centralized data and struggle to cope with the decentralized and diverse nature of federated medical settings. In this work, we present MedFedPure, a personalized federated learning defense framework designed to protect diagnostic AI models at inference time without compromising privacy or accuracy. MedFedPure combines three key elements: (1) a personalized FL model that adapts to the unique data distribution of each institution; (2) a Masked Autoencoder (MAE) that detects suspicious inputs by exposing hidden perturbations; and (3) an adaptive diffusion-based purification module that selectively cleans only the flagged scans before classification. Together, these steps offer robust protection while preserving the integrity of normal, benign images. We evaluated MedFedPure on the Br35H brain MRI dataset. The results show a significant gain in adversarial robustness, improving performance from 49.50% to 87.33% under strong attacks, while maintaining a high clean accuracy of 97.67%. By operating locally and in real time during diagnosis, our framework provides a practical path to deploying secure, trustworthy, and privacy-preserving AI tools in clinical workflows. Index Terms: cancer, tumor detection, federated learning, masked autoencoder, diffusion, privacy
RRTS Dataset: A Benchmark Colonoscopy Dataset from Resource-Limited Settings for Computer-Aided Diagnosis Research
Nov 10, 2025Background and Objective: Colorectal cancer prevention relies on early detection of polyps during colonoscopy. Existing public datasets, such as CVC-ClinicDB and Kvasir-SEG, provide valuable benchmarks but are limited by small sample sizes, curated image selection, or lack of real-world artifacts. There remains a need for datasets that capture the complexity of clinical practice, particularly in resource-constrained settings. Methods: We introduce a dataset, BUET Polyp Dataset (BPD), of colonoscopy images collected using Olympus 170 and Pentax i-Scan series endoscopes under routine clinical conditions. The dataset contains images with corresponding expert-annotated binary masks, reflecting diverse challenges such as motion blur, specular highlights, stool artifacts, blood, and low-light frames. Annotations were manually reviewed by clinical experts to ensure quality. To demonstrate baseline performance, we provide benchmark results for classification using VGG16, ResNet50, and InceptionV3, and for segmentation using UNet variants with VGG16, ResNet34, and InceptionV4 backbones. Results: The dataset comprises 1,288 images with polyps from 164 patients with corresponding ground-truth masks and 1,657 polyp-free images from 31 patients. Benchmarking experiments achieved up to 90.8% accuracy for binary classification (VGG16) and a maximum Dice score of 0.64 with InceptionV4-UNet for segmentation. Performance was lower compared to curated datasets, reflecting the real-world difficulty of images with artifacts and variable quality.
PSO-XAI: A PSO-Enhanced Explainable AI Framework for Reliable Breast Cancer Detection
Oct 23, 2025Breast cancer is considered the most critical and frequently diagnosed cancer in women worldwide, leading to an increase in cancer-related mortality. Early and accurate detection is crucial as it can help mitigate possible threats while improving survival rates. In terms of prediction, conventional diagnostic methods are often limited by variability, cost, and, most importantly, risk of misdiagnosis. To address these challenges, machine learning (ML) has emerged as a powerful tool for computer-aided diagnosis, with feature selection playing a vital role in improving model performance and interpretability. This research study proposes an integrated framework that incorporates customized Particle Swarm Optimization (PSO) for feature selection. This framework has been evaluated on a comprehensive set of 29 different models, spanning classical classifiers, ensemble techniques, neural networks, probabilistic algorithms, and instance-based algorithms. To ensure interpretability and clinical relevance, the study uses cross-validation in conjunction with explainable AI methods. Experimental evaluation showed that the proposed approach achieved a superior score of 99.1\% across all performance metrics, including accuracy and precision, while effectively reducing dimensionality and providing transparent, model-agnostic explanations. The results highlight the potential of combining swarm intelligence with explainable ML for robust, trustworthy, and clinically meaningful breast cancer diagnosis.
Dark-Field X-Ray Imaging Significantly Improves Deep-Learning based Detection of Synthetic Early-Stage Lung Tumors in Preclinical Models
Oct 31, 2025Low-dose computed tomography (LDCT) is the current standard for lung cancer screening, yet its adoption and accessibility remain limited. Many regions lack LDCT infrastructure, and even among those screened, early-stage cancer detection often yield false positives, as shown in the National Lung Screening Trial (NLST) with a sensitivity of 93.8 percent and a false-positive rate of 26.6 percent. We aim to investigate whether X-ray dark-field imaging (DFI) radiograph, a technique sensitive to small-angle scatter from alveolar microstructure and less susceptible to organ shadowing, can significantly improve early-stage lung tumor detection when coupled with deep-learning segmentation. Using paired attenuation (ATTN) and DFI radiograph images of euthanized mouse lungs, we generated realistic synthetic tumors with irregular boundaries and intensity profiles consistent with physical lung contrast. A U-Net segmentation network was trained on small patches using either ATTN, DFI, or a combination of ATTN and DFI channels. Results show that the DFI-only model achieved a true-positive detection rate of 83.7 percent, compared with 51 percent for ATTN-only, while maintaining comparable specificity (90.5 versus 92.9 percent). The combined ATTN and DFI input achieved 79.6 percent sensitivity and 97.6 percent specificity. In conclusion, DFI substantially improves early-tumor detectability in comparison to standard attenuation radiography and shows potential as an accessible, low-cost, low-dose alternative for pre-clinical or limited-resource screening where LDCT is unavailable.
MV-MLM: Bridging Multi-View Mammography and Language for Breast Cancer Diagnosis and Risk Prediction
Oct 30, 2025



Large annotated datasets are essential for training robust Computer-Aided Diagnosis (CAD) models for breast cancer detection or risk prediction. However, acquiring such datasets with fine-detailed annotation is both costly and time-consuming. Vision-Language Models (VLMs), such as CLIP, which are pre-trained on large image-text pairs, offer a promising solution by enhancing robustness and data efficiency in medical imaging tasks. This paper introduces a novel Multi-View Mammography and Language Model for breast cancer classification and risk prediction, trained on a dataset of paired mammogram images and synthetic radiology reports. Our MV-MLM leverages multi-view supervision to learn rich representations from extensive radiology data by employing cross-modal self-supervision across image-text pairs. This includes multiple views and the corresponding pseudo-radiology reports. We propose a novel joint visual-textual learning strategy to enhance generalization and accuracy performance over different data types and tasks to distinguish breast tissues or cancer characteristics(calcification, mass) and utilize these patterns to understand mammography images and predict cancer risk. We evaluated our method on both private and publicly available datasets, demonstrating that the proposed model achieves state-of-the-art performance in three classification tasks: (1) malignancy classification, (2) subtype classification, and (3) image-based cancer risk prediction. Furthermore, the model exhibits strong data efficiency, outperforming existing fully supervised or VLM baselines while trained on synthetic text reports and without the need for actual radiology reports.
Histology-informed tiling of whole tissue sections improves the interpretability and predictability of cancer relapse and genetic alterations
Nov 13, 2025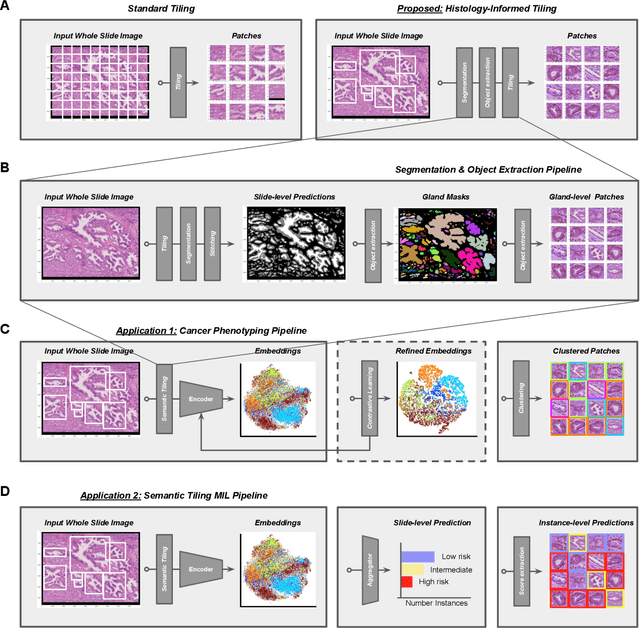
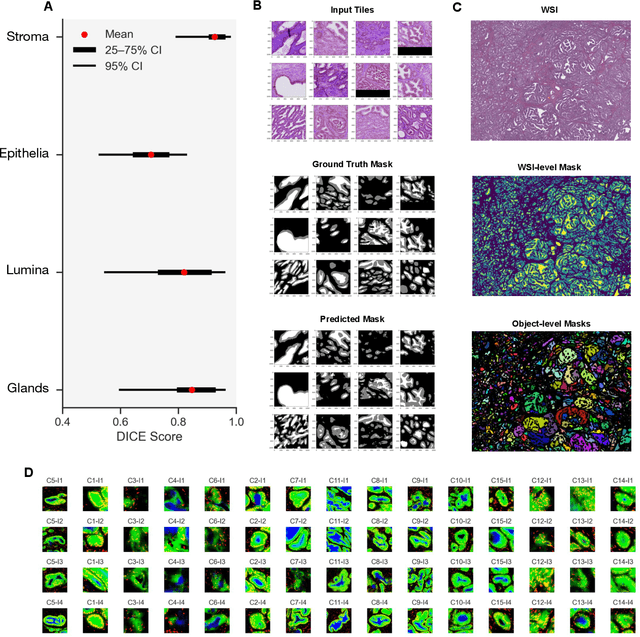
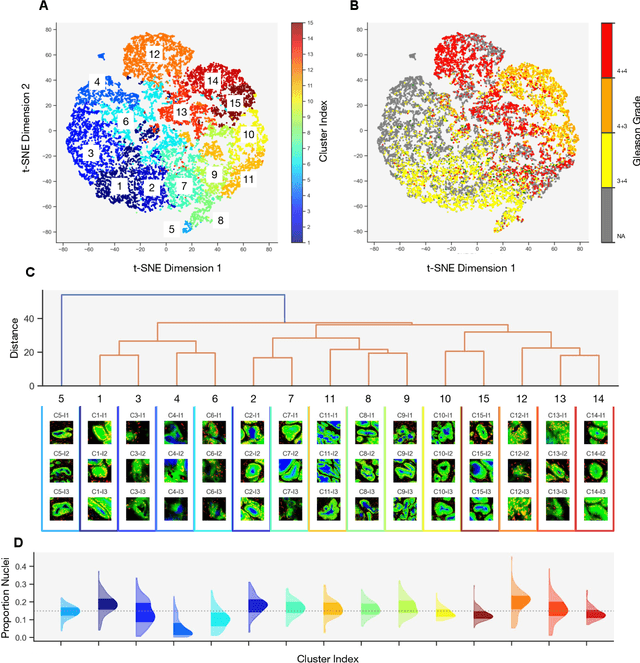
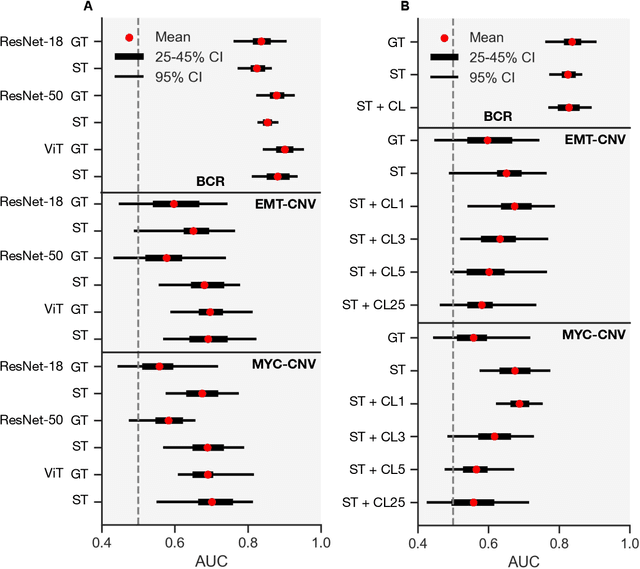
Histopathologists establish cancer grade by assessing histological structures, such as glands in prostate cancer. Yet, digital pathology pipelines often rely on grid-based tiling that ignores tissue architecture. This introduces irrelevant information and limits interpretability. We introduce histology-informed tiling (HIT), which uses semantic segmentation to extract glands from whole slide images (WSIs) as biologically meaningful input patches for multiple-instance learning (MIL) and phenotyping. Trained on 137 samples from the ProMPT cohort, HIT achieved a gland-level Dice score of 0.83 +/- 0.17. By extracting 380,000 glands from 760 WSIs across ICGC-C and TCGA-PRAD cohorts, HIT improved MIL models AUCs by 10% for detecting copy number variation (CNVs) in genes related to epithelial-mesenchymal transitions (EMT) and MYC, and revealed 15 gland clusters, several of which were associated with cancer relapse, oncogenic mutations, and high Gleason. Therefore, HIT improved the accuracy and interpretability of MIL predictions, while streamlining computations by focussing on biologically meaningful structures during feature extraction.
Rank-Aware Agglomeration of Foundation Models for Immunohistochemistry Image Cell Counting
Nov 16, 2025Accurate cell counting in immunohistochemistry (IHC) images is critical for quantifying protein expression and aiding cancer diagnosis. However, the task remains challenging due to the chromogen overlap, variable biomarker staining, and diverse cellular morphologies. Regression-based counting methods offer advantages over detection-based ones in handling overlapped cells, yet rarely support end-to-end multi-class counting. Moreover, the potential of foundation models remains largely underexplored in this paradigm. To address these limitations, we propose a rank-aware agglomeration framework that selectively distills knowledge from multiple strong foundation models, leveraging their complementary representations to handle IHC heterogeneity and obtain a compact yet effective student model, CountIHC. Unlike prior task-agnostic agglomeration strategies that either treat all teachers equally or rely on feature similarity, we design a Rank-Aware Teacher Selecting (RATS) strategy that models global-to-local patch rankings to assess each teacher's inherent counting capacity and enable sample-wise teacher selection. For multi-class cell counting, we introduce a fine-tuning stage that reformulates the task as vision-language alignment. Discrete semantic anchors derived from structured text prompts encode both category and quantity information, guiding the regression of class-specific density maps and improving counting for overlapping cells. Extensive experiments demonstrate that CountIHC surpasses state-of-the-art methods across 12 IHC biomarkers and 5 tissue types, while exhibiting high agreement with pathologists' assessments. Its effectiveness on H&E-stained data further confirms the scalability of the proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge