Zhouwu Liu
Decoding Translation-Related Functional Sequences in 5'UTRs Using Interpretable Deep Learning Models
Jul 22, 2025Abstract:Understanding how 5' untranslated regions (5'UTRs) regulate mRNA translation is critical for controlling protein expression and designing effective therapeutic mRNAs. While recent deep learning models have shown promise in predicting translational efficiency from 5'UTR sequences, most are constrained by fixed input lengths and limited interpretability. We introduce UTR-STCNet, a Transformer-based architecture for flexible and biologically grounded modeling of variable-length 5'UTRs. UTR-STCNet integrates a Saliency-Aware Token Clustering (SATC) module that iteratively aggregates nucleotide tokens into multi-scale, semantically meaningful units based on saliency scores. A Saliency-Guided Transformer (SGT) block then captures both local and distal regulatory dependencies using a lightweight attention mechanism. This combined architecture achieves efficient and interpretable modeling without input truncation or increased computational cost. Evaluated across three benchmark datasets, UTR-STCNet consistently outperforms state-of-the-art baselines in predicting mean ribosome load (MRL), a key proxy for translational efficiency. Moreover, the model recovers known functional elements such as upstream AUGs and Kozak motifs, highlighting its potential for mechanistic insight into translation regulation.
Task-oriented Self-supervised Learning for Anomaly Detection in Electroencephalography
Jul 04, 2022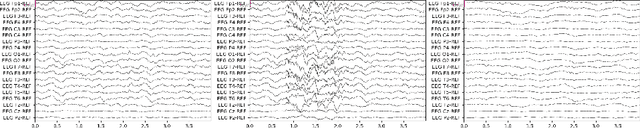
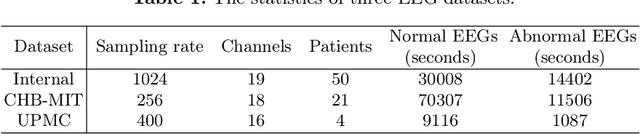
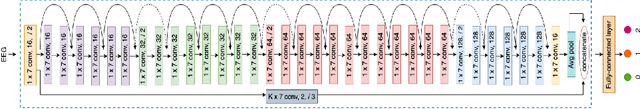
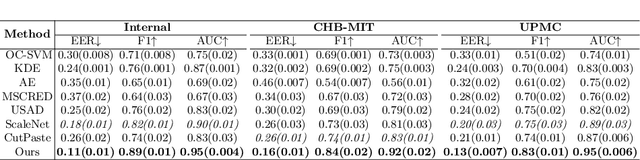
Abstract:Accurate automated analysis of electroencephalography (EEG) would largely help clinicians effectively monitor and diagnose patients with various brain diseases. Compared to supervised learning with labelled disease EEG data which can train a model to analyze specific diseases but would fail to monitor previously unseen statuses, anomaly detection based on only normal EEGs can detect any potential anomaly in new EEGs. Different from existing anomaly detection strategies which do not consider any property of unavailable abnormal data during model development, a task-oriented self-supervised learning approach is proposed here which makes use of available normal EEGs and expert knowledge about abnormal EEGs to train a more effective feature extractor for the subsequent development of anomaly detector. In addition, a specific two branch convolutional neural network with larger kernels is designed as the feature extractor such that it can more easily extract both larger scale and small-scale features which often appear in unavailable abnormal EEGs. The effectively designed and trained feature extractor has shown to be able to extract better feature representations from EEGs for development of anomaly detector based on normal data and future anomaly detection for new EEGs, as demonstrated on three EEG datasets. The code is available at https://github.com/ironing/EEG-AD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge