Zhonghao Liu
Automated glenoid bone loss measurement and segmentation in CT scans for pre-operative planning in shoulder instability
Nov 18, 2025Abstract:Reliable measurement of glenoid bone loss is essential for operative planning in shoulder instability, but current manual and semi-automated methods are time-consuming and often subject to interreader variability. We developed and validated a fully automated deep learning pipeline for measuring glenoid bone loss on three-dimensional computed tomography (CT) scans using a linear-based, en-face view, best-circle method. Shoulder CT images of 91 patients (average age, 40 years; range, 14-89 years; 65 men) were retrospectively collected along with manual labels including glenoid segmentation, landmarks, and bone loss measurements. The multi-stage algorithm has three main stages: (1) segmentation, where we developed a U-Net to automatically segment the glenoid and humerus; (2) anatomical landmark detection, where a second network predicts glenoid rim points; and (3) geometric fitting, where we applied principal component analysis (PCA), projection, and circle fitting to compute the percentage of bone loss. The automated measurements showed strong agreement with consensus readings and exceeded surgeon-to-surgeon consistency (intraclass correlation coefficient (ICC) 0.84 vs 0.78), including in low- and high-bone-loss subgroups (ICC 0.71 vs 0.63 and 0.83 vs 0.21, respectively; P < 0.001). For classifying patients into low, medium, and high bone-loss categories, the pipeline achieved a recall of 0.714 for low and 0.857 for high severity, with no low cases misclassified as high or vice versa. These results suggest that our method is a time-efficient and clinically reliable tool for preoperative planning in shoulder instability and for screening patients with substantial glenoid bone loss. Code and dataset are available at https://github.com/Edenliu1/Auto-Glenoid-Measurement-DL-Pipeline.
A Deep Learning Algorithm for One-step Contour Aware Nuclei Segmentation of Histopathological Images
Mar 07, 2018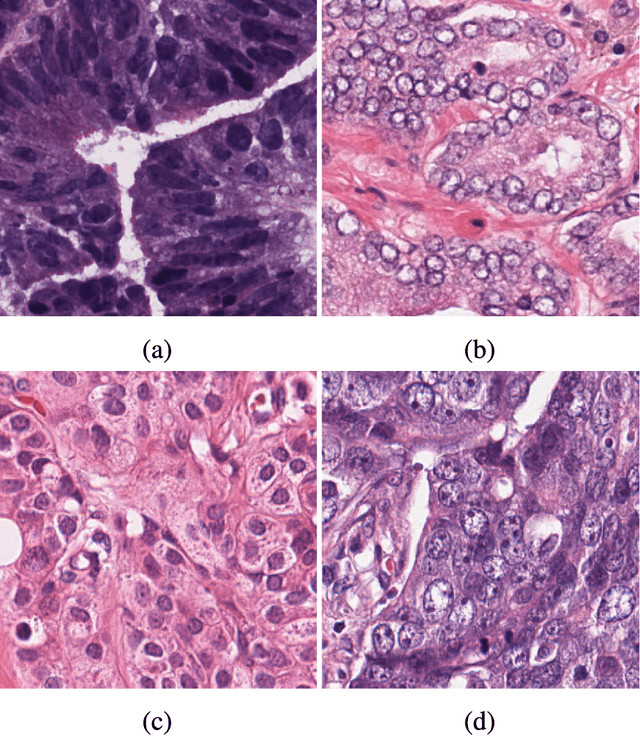
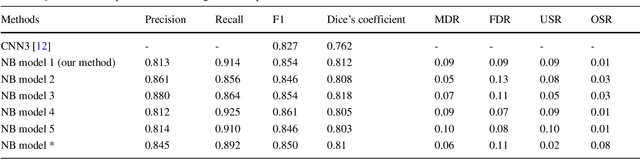
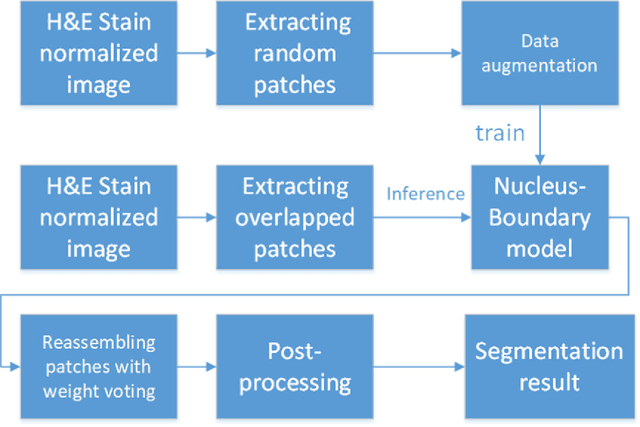
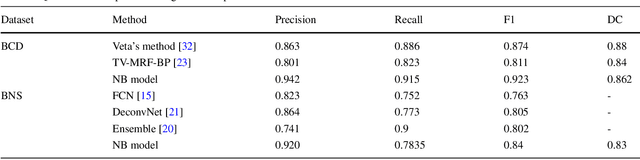
Abstract:This paper addresses the task of nuclei segmentation in high-resolution histopathological images. We propose an auto- matic end-to-end deep neural network algorithm for segmenta- tion of individual nuclei. A nucleus-boundary model is introduced to predict nuclei and their boundaries simultaneously using a fully convolutional neural network. Given a color normalized image, the model directly outputs an estimated nuclei map and a boundary map. A simple, fast and parameter-free post-processing procedure is performed on the estimated nuclei map to produce the final segmented nuclei. An overlapped patch extraction and assembling method is also designed for seamless prediction of nuclei in large whole-slide images. We also show the effectiveness of data augmentation methods for nuclei segmentation task. Our experiments showed our method outperforms prior state-of-the- art methods. Moreover, it is efficient that one 1000X1000 image can be segmented in less than 5 seconds. This makes it possible to precisely segment the whole-slide image in acceptable time
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge