Zhendi Gong
SSL-MedSAM2: A Semi-supervised Medical Image Segmentation Framework Powered by Few-shot Learning of SAM2
Dec 12, 2025Abstract:Despite the success of deep learning based models in medical image segmentation, most state-of-the-art (SOTA) methods perform fully-supervised learning, which commonly rely on large scale annotated training datasets. However, medical image annotation is highly time-consuming, hindering its clinical applications. Semi-supervised learning (SSL) has been emerged as an appealing strategy in training with limited annotations, largely reducing the labelling cost. We propose a novel SSL framework SSL-MedSAM2, which contains a training-free few-shot learning branch TFFS-MedSAM2 based on the pretrained large foundation model Segment Anything Model 2 (SAM2) for pseudo label generation, and an iterative fully-supervised learning branch FSL-nnUNet based on nnUNet for pseudo label refinement. The results on MICCAI2025 challenge CARE-LiSeg (Liver Segmentation) demonstrate an outstanding performance of SSL-MedSAM2 among other methods. The average dice scores on the test set in GED4 and T1 MRI are 0.9710 and 0.9648 respectively, and the Hausdorff distances are 20.07 and 21.97 respectively. The code is available via https://github.com/naisops/SSL-MedSAM2/tree/main.
MO-CTranS: A unified multi-organ segmentation model learning from multiple heterogeneously labelled datasets
Mar 28, 2025


Abstract:Multi-organ segmentation holds paramount significance in many clinical tasks. In practice, compared to large fully annotated datasets, multiple small datasets are often more accessible and organs are not labelled consistently. Normally, an individual model is trained for each of these datasets, which is not an effective way of using data for model learning. It remains challenging to train a single model that can robustly learn from several partially labelled datasets due to label conflict and data imbalance problems. We propose MO-CTranS: a single model that can overcome such problems. MO-CTranS contains a CNN-based encoder and a Transformer-based decoder, which are connected in a multi-resolution manner. Task-specific tokens are introduced in the decoder to help differentiate label discrepancies. Our method was evaluated and compared to several baseline models and state-of-the-art (SOTA) solutions on abdominal MRI datasets that were acquired in different views (i.e. axial and coronal) and annotated for different organs (i.e. liver, kidney, spleen). Our method achieved better performance (most were statistically significant) than the compared methods. Github link: https://github.com/naisops/MO-CTranS.
ConvTransSeg: A Multi-resolution Convolution-Transformer Network for Medical Image Segmentation
Oct 13, 2022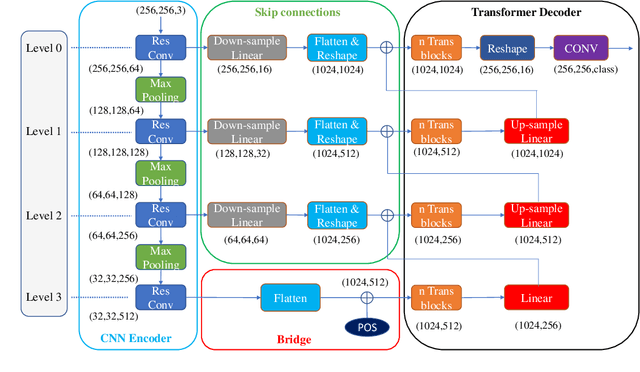
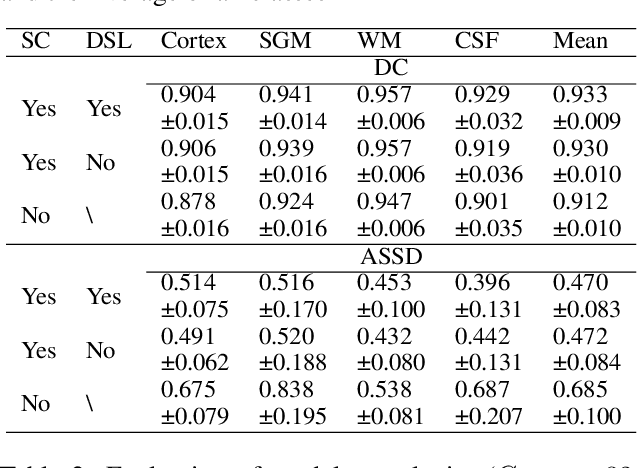
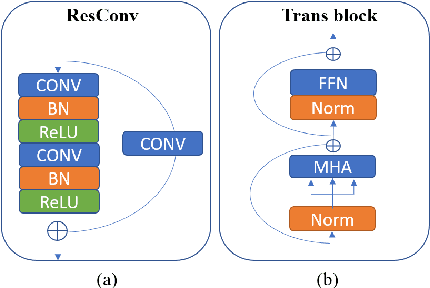
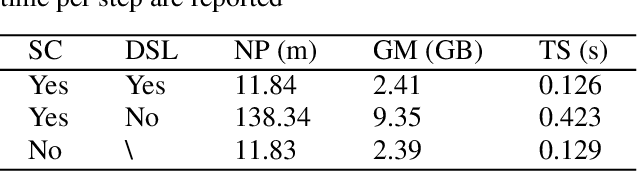
Abstract:Convolutional neural networks (CNNs) achieved the state-of-the-art performance in medical image segmentation due to their ability to extract highly complex feature representations. However, it is argued in recent studies that traditional CNNs lack the intelligence to capture long-term dependencies of different image regions. Following the success of applying Transformer models on natural language processing tasks, the medical image segmentation field has also witnessed growing interest in utilizing Transformers, due to their ability to capture long-range contextual information. However, unlike CNNs, Transformers lack the ability to learn local feature representations. Thus, to fully utilize the advantages of both CNNs and Transformers, we propose a hybrid encoder-decoder segmentation model (ConvTransSeg). It consists of a multi-layer CNN as the encoder for feature learning and the corresponding multi-level Transformer as the decoder for segmentation prediction. The encoder and decoder are interconnected in a multi-resolution manner. We compared our method with many other state-of-the-art hybrid CNN and Transformer segmentation models on binary and multiple class image segmentation tasks using several public medical image datasets, including skin lesion, polyp, cell and brain tissue. The experimental results show that our method achieves overall the best performance in terms of Dice coefficient and average symmetric surface distance measures with low model complexity and memory consumption. In contrast to most Transformer-based methods that we compared, our method does not require the use of pre-trained models to achieve similar or better performance. The code is freely available for research purposes on Github: (the link will be added upon acceptance).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge