Zaifa Xue
Cooperative Causal GraphSAGE
May 20, 2025Abstract:GraphSAGE is a widely used graph neural network. The introduction of causal inference has improved its robust performance and named as Causal GraphSAGE. However, Causal GraphSAGE focuses on measuring causal weighting among individual nodes, but neglecting the cooperative relationships among sampling nodes as a whole. To address this issue, this paper proposes Cooperative Causal GraphSAGE (CoCa-GraphSAGE), which combines cooperative game theory with Causal GraphSAGE. Initially, a cooperative causal structure model is constructed in the case of cooperation based on the graph structure. Subsequently, Cooperative Causal sampling (CoCa-sampling) algorithm is proposed, employing the Shapley values to calculate the cooperative contribution based on causal weights of the nodes sets. CoCa-sampling guides the selection of nodes with significant cooperative causal effects during the neighborhood sampling process, thus integrating the selected neighborhood features under cooperative relationships, which takes the sampled nodes as a whole and generates more stable target node embeddings. Experiments on publicly available datasets show that the proposed method has comparable classification performance to the compared methods and outperforms under perturbations, demonstrating the robustness improvement by CoCa-sampling.
Patient-Specific Game-Based Transfer Method for Parkinson's Disease Severity Prediction
Aug 12, 2022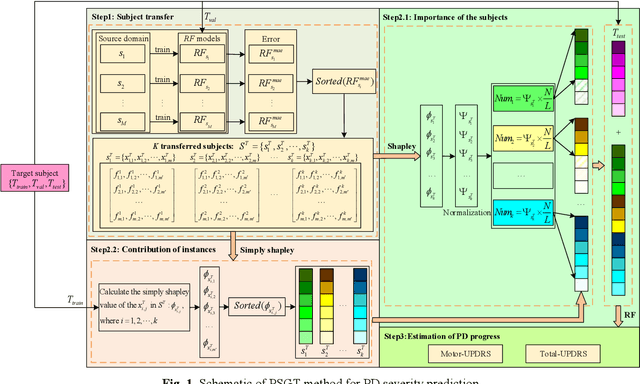
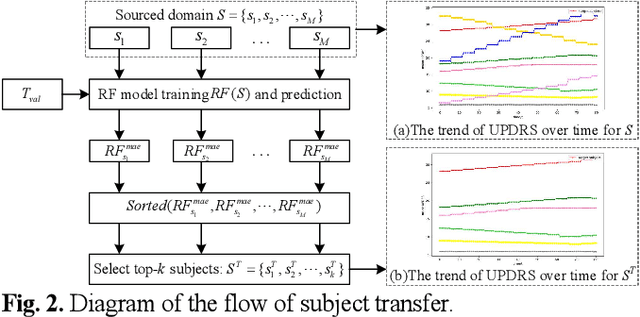


Abstract:Dysphonia is one of the early symptoms of Parkinson's disease (PD). Most existing methods use feature selection methods to find the optimal subset of voice features for all PD patients. Few have considered the heterogeneity between patients, which implies the need to provide specific prediction models for different patients. However, building the specific model faces the challenge of small sample size, which makes it lack generalization ability. Instance transfer is an effective way to solve this problem. Therefore, this paper proposes a patient-specific game-based transfer (PSGT) method for PD severity prediction. First, a selection mechanism is used to select PD patients with similar disease trends to the target patient from the source domain, which greatly reduces the risk of negative transfer. Then, the contribution of the transferred subjects and their instances to the disease estimation of the target subject is fairly evaluated by the Shapley value, which improves the interpretability of the method. Next, the proportion of valid instances in the transferred subjects is determined, and the instances with higher contribution are transferred to further reduce the difference between the transferred instance subset and the target subject. Finally, the selected subset of instances is added to the training set of the target subject, and the extended data is fed into the random forest to improve the performance of the method. Parkinson's telemonitoring dataset is used to evaluate the feasibility and effectiveness. Experiment results show that the PSGT has better performance in both prediction error and stability over compared methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge