Yinping Ma
MixGRPO: Unlocking Flow-based GRPO Efficiency with Mixed ODE-SDE
Jul 29, 2025Abstract:Although GRPO substantially enhances flow matching models in human preference alignment of image generation, methods such as FlowGRPO still exhibit inefficiency due to the necessity of sampling and optimizing over all denoising steps specified by the Markov Decision Process (MDP). In this paper, we propose $\textbf{MixGRPO}$, a novel framework that leverages the flexibility of mixed sampling strategies through the integration of stochastic differential equations (SDE) and ordinary differential equations (ODE). This streamlines the optimization process within the MDP to improve efficiency and boost performance. Specifically, MixGRPO introduces a sliding window mechanism, using SDE sampling and GRPO-guided optimization only within the window, while applying ODE sampling outside. This design confines sampling randomness to the time-steps within the window, thereby reducing the optimization overhead, and allowing for more focused gradient updates to accelerate convergence. Additionally, as time-steps beyond the sliding window are not involved in optimization, higher-order solvers are supported for sampling. So we present a faster variant, termed $\textbf{MixGRPO-Flash}$, which further improves training efficiency while achieving comparable performance. MixGRPO exhibits substantial gains across multiple dimensions of human preference alignment, outperforming DanceGRPO in both effectiveness and efficiency, with nearly 50% lower training time. Notably, MixGRPO-Flash further reduces training time by 71%. Codes and models are available at $\href{https://github.com/Tencent-Hunyuan/MixGRPO}{MixGRPO}$.
Deep manifold learning reveals hidden dynamics of proteasome autoregulation
Dec 23, 2020

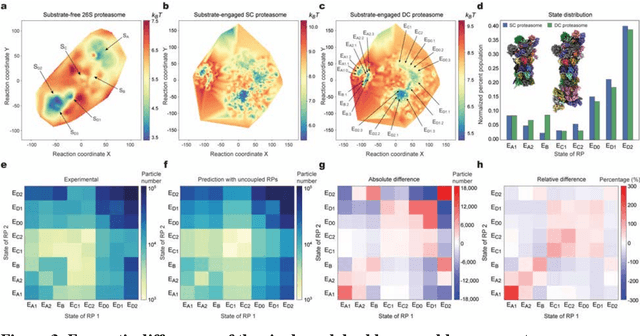

Abstract:The 2.5-MDa 26S proteasome maintains proteostasis and regulates myriad cellular processes. How polyubiquitylated substrate interactions regulate proteasome activity is not understood. Here we introduce a deep manifold learning framework, named AlphaCryo4D, which enables atomic-level cryogenic electron microscopy (cryo-EM) reconstructions of nonequilibrium conformational continuum and reconstitutes hidden dynamics of proteasome autoregulation in the act of substrate degradation. AlphaCryo4D integrates 3D deep residual learning with manifold embedding of free-energy landscapes, which directs 3D clustering via an energy-based particle-voting algorithm. In blind assessments using simulated heterogeneous cryo-EM datasets, AlphaCryo4D achieved 3D classification accuracy three times that of conventional method and reconstructed continuous conformational changes of a 130-kDa protein at sub-3-angstrom resolution. By using AlphaCryo4D to analyze a single experimental cryo-EM dataset, we identified 64 conformers of the substrate-bound human 26S proteasome, revealing conformational entanglement of two regulatory particles in the doubly capped holoenzymes and their energetic differences with singly capped ones. Novel ubiquitin-binding sites are discovered on the RPN2, RPN10 and Alpha5 subunits to remodel polyubiquitin chains for deubiquitylation and recycle. Importantly, AlphaCryo4D choreographs single-nucleotide-exchange dynamics of proteasomal AAA-ATPase motor during translocation initiation, which upregulates proteolytic activity by allosterically promoting nucleophilic attack. Our systemic analysis illuminates a grand hierarchical allostery for proteasome autoregulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge