Yanke Li
KarmaTS: A Universal Simulation Platform for Multivariate Time Series with Functional Causal Dynamics
Nov 14, 2025Abstract:We introduce KarmaTS, an interactive framework for constructing lag-indexed, executable spatiotemporal causal graphical models for multivariate time series (MTS) simulation. Motivated by the challenge of access-restricted physiological data, KarmaTS generates synthetic MTS with known causal dynamics and augments real-world datasets with expert knowledge. The system constructs a discrete-time structural causal process (DSCP) by combining expert knowledge and algorithmic proposals in a mixed-initiative, human-in-the-loop workflow. The resulting DSCP supports simulation and causal interventions, including those under user-specified distribution shifts. KarmaTS handles mixed variable types, contemporaneous and lagged edges, and modular edge functionals ranging from parameterizable templates to neural network models. Together, these features enable flexible validation and benchmarking of causal discovery algorithms through expert-informed simulation.
DAPDAG: Domain Adaptation via Perturbed DAG Reconstruction
Aug 02, 2022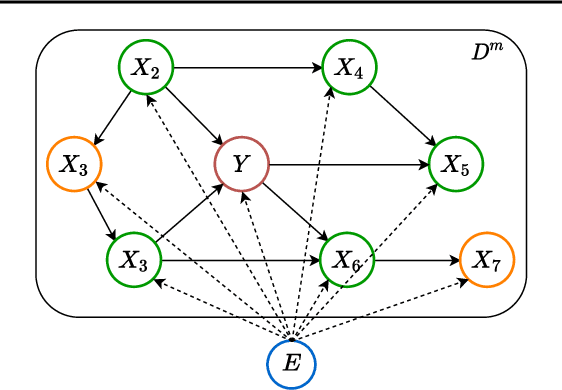
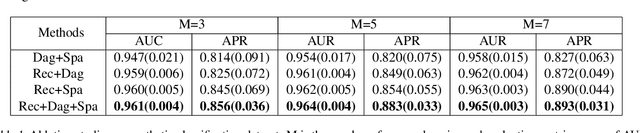
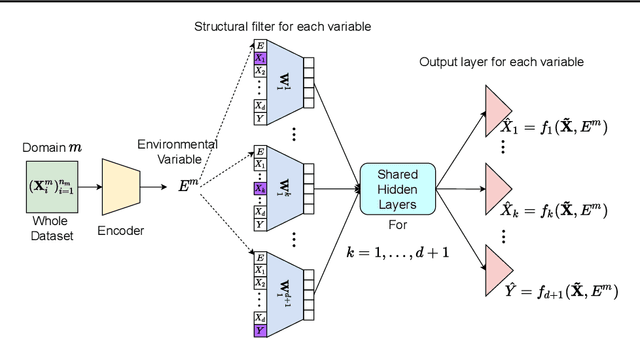
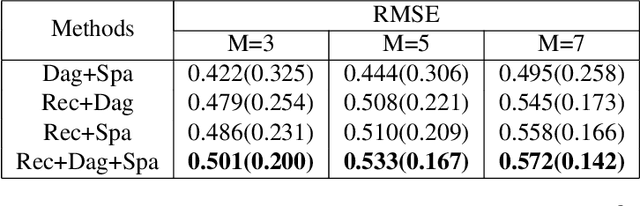
Abstract:Leveraging labelled data from multiple domains to enable prediction in another domain without labels is a significant, yet challenging problem. To address this problem, we introduce the framework DAPDAG (\textbf{D}omain \textbf{A}daptation via \textbf{P}erturbed \textbf{DAG} Reconstruction) and propose to learn an auto-encoder that undertakes inference on population statistics given features and reconstructing a directed acyclic graph (DAG) as an auxiliary task. The underlying DAG structure is assumed invariant among observed variables whose conditional distributions are allowed to vary across domains led by a latent environmental variable $E$. The encoder is designed to serve as an inference device on $E$ while the decoder reconstructs each observed variable conditioned on its graphical parents in the DAG and the inferred $E$. We train the encoder and decoder jointly in an end-to-end manner and conduct experiments on synthetic and real datasets with mixed variables. Empirical results demonstrate that reconstructing the DAG benefits the approximate inference. Furthermore, our approach can achieve competitive performance against other benchmarks in prediction tasks, with better adaptation ability, especially in the target domain significantly different from the source domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge