Vivian Hui
Exploration of Augmentation Strategies in Multi-modal Retrieval-Augmented Generation for the Biomedical Domain: A Case Study Evaluating Question Answering in Glycobiology
Dec 18, 2025Abstract:Multi-modal retrieval-augmented generation (MM-RAG) promises grounded biomedical QA, but it is unclear when to (i) convert figures/tables into text versus (ii) use optical character recognition (OCR)-free visual retrieval that returns page images and leaves interpretation to the generator. We study this trade-off in glycobiology, a visually dense domain. We built a benchmark of 120 multiple-choice questions (MCQs) from 25 papers, stratified by retrieval difficulty (easy text, medium figures/tables, hard cross-evidence). We implemented four augmentations-None, Text RAG, Multi-modal conversion, and late-interaction visual retrieval (ColPali)-using Docling parsing and Qdrant indexing. We evaluated mid-size open-source and frontier proprietary models (e.g., Gemma-3-27B-IT, GPT-4o family). Additional testing used the GPT-5 family and multiple visual retrievers (ColPali/ColQwen/ColFlor). Accuracy with Agresti-Coull 95% confidence intervals (CIs) was computed over 5 runs per configuration. With Gemma-3-27B-IT, Text and Multi-modal augmentation outperformed OCR-free retrieval (0.722-0.740 vs. 0.510 average accuracy). With GPT-4o, Multi-modal achieved 0.808, with Text 0.782 and ColPali 0.745 close behind; within-model differences were small. In follow-on experiments with the GPT-5 family, the best results with ColPali and ColFlor improved by ~2% to 0.828 in both cases. In general, across the GPT-5 family, ColPali, ColQwen, and ColFlor were statistically indistinguishable. GPT-5-nano trailed larger GPT-5 variants by roughly 8-10%. Pipeline choice is capacity-dependent: converting visuals to text lowers the reader burden and is more reliable for mid-size models, whereas OCR-free visual retrieval becomes competitive under frontier models. Among retrievers, ColFlor offers parity with heavier options at a smaller footprint, making it an efficient default when strong generators are available.
Privacy Challenges and Solutions in Retrieval-Augmented Generation-Enhanced LLMs for Healthcare Chatbots: A Review of Applications, Risks, and Future Directions
Nov 17, 2025Abstract:Retrieval-augmented generation (RAG) has rapidly emerged as a transformative approach for integrating large language models into clinical and biomedical workflows. However, privacy risks, such as protected health information (PHI) exposure, remain inconsistently mitigated. This review provides a thorough analysis of the current landscape of RAG applications in healthcare, including (i) sensitive data type across clinical scenarios, (ii) the associated privacy risks, (iii) current and emerging data-privacy protection mechanisms and (iv) future direction for patient data privacy protection. We synthesize 23 articles on RAG applications in healthcare and systematically analyze privacy challenges through a pipeline-structured framework encompassing data storage, transmission, retrieval and generation stages, delineating potential failure modes, their underlying causes in threat models and system mechanisms, and their practical implications. Building on this analysis, we critically review 17 articles on privacy-preserving strategies for RAG systems. Our evaluation reveals critical gaps, including insufficient clinical validation, absence of standardized evaluation frameworks, and lack of automated assessment tools. We propose actionable directions based on these limitations and conclude with a call to action. This review provides researchers and practitioners with a structured framework for understanding privacy vulnerabilities in healthcare RAG and offers a roadmap toward developing systems that achieve both clinical effectiveness and robust privacy preservation.
Toward Improving Health Literacy in Patient Education Materials with Neural Machine Translation Models
Sep 14, 2022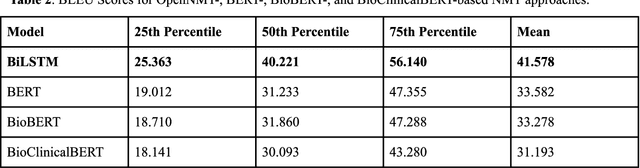
Abstract:Health literacy is the central focus of Healthy People 2030, the fifth iteration of the U.S. national goals and objectives. People with low health literacy usually have trouble understanding health information, following post-visit instructions, and using prescriptions, which results in worse health outcomes and serious health disparities. In this study, we propose to leverage natural language processing techniques to improve health literacy in patient education materials by automatically translating illiterate languages in a given sentence. We scraped patient education materials from four online health information websites: MedlinePlus.gov, Drugs.com, Mayoclinic.org and Reddit.com. We trained and tested the state-of-the-art neural machine translation (NMT) models on a silver standard training dataset and a gold standard testing dataset, respectively. The experimental results showed that the Bidirectional Long Short-Term Memory (BiLSTM) NMT model outperformed Bidirectional Encoder Representations from Transformers (BERT)-based NMT models. We also verified the effectiveness of NMT models in translating health illiterate languages by comparing the ratio of health illiterate language in the sentence. The proposed NMT models were able to identify the correct complicated words and simplify into layman language while at the same time the models suffer from sentence completeness, fluency, readability, and have difficulty in translating certain medical terms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge