Vivek Kurien George
Advancing Neural Network Performance through Emergence-Promoting Initialization Scheme
Jul 26, 2024Abstract:We introduce a novel yet straightforward neural network initialization scheme that modifies conventional methods like Xavier and Kaiming initialization. Inspired by the concept of emergence and leveraging the emergence measures proposed by Li (2023), our method adjusts the layer-wise weight scaling factors to achieve higher emergence values. This enhancement is easy to implement, requiring no additional optimization steps for initialization compared to GradInit. We evaluate our approach across various architectures, including MLP and convolutional architectures for image recognition, and transformers for machine translation. We demonstrate substantial improvements in both model accuracy and training speed, with and without batch normalization. The simplicity, theoretical innovation, and demonstrable empirical advantages of our method make it a potent enhancement to neural network initialization practices. These results suggest a promising direction for leveraging emergence to improve neural network training methodologies. Code is available at: https://github.com/johnnyjingzeli/EmergenceInit.
Learning without gradient descent encoded by the dynamics of a neurobiological model
Mar 23, 2021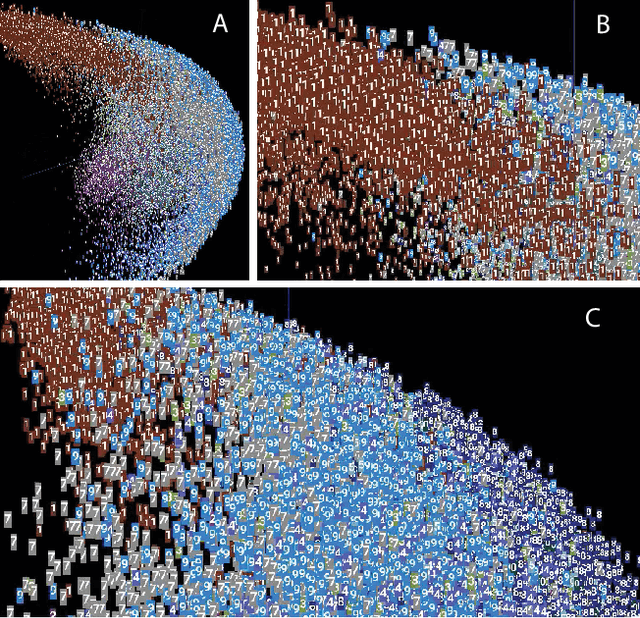
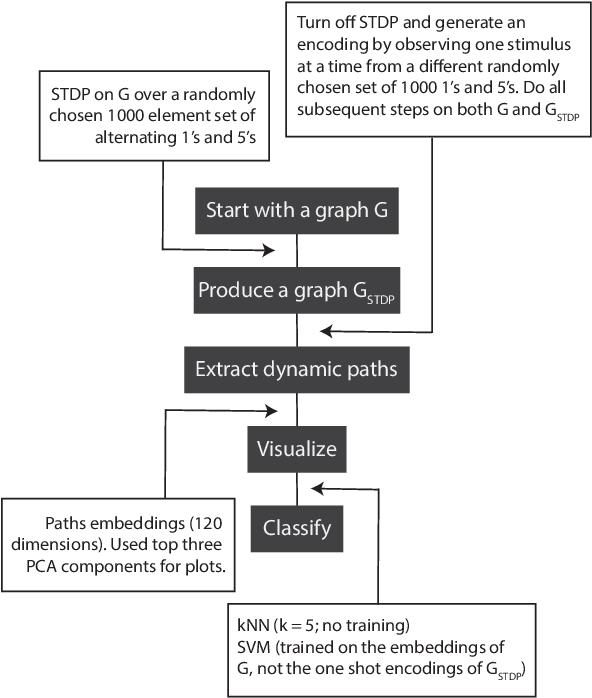
Abstract:The success of state-of-the-art machine learning is essentially all based on different variations of gradient descent algorithms that minimize some version of a cost or loss function. A fundamental limitation, however, is the need to train these systems in either supervised or unsupervised ways by exposing them to typically large numbers of training examples. Here, we introduce a fundamentally novel conceptual approach to machine learning that takes advantage of a neurobiologically derived model of dynamic signaling, constrained by the geometric structure of a network. We show that MNIST images can be uniquely encoded and classified by the dynamics of geometric networks with nearly state-of-the-art accuracy in an unsupervised way, and without the need for any training.
Generalizable Machine Learning in Neuroscience using Graph Neural Networks
Oct 16, 2020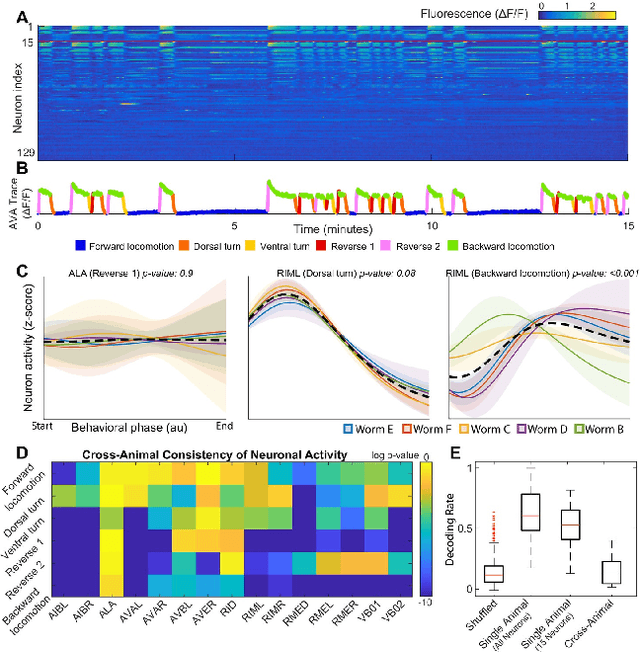
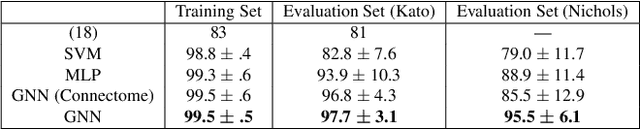
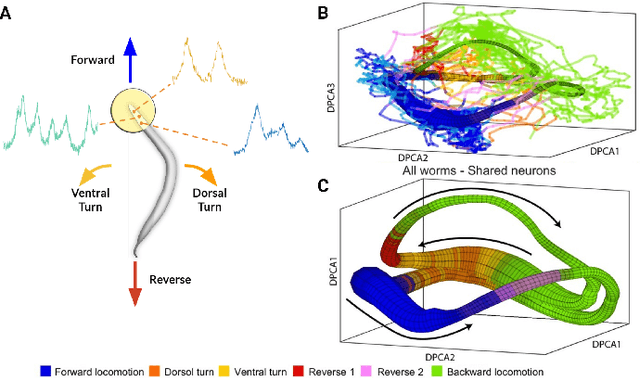
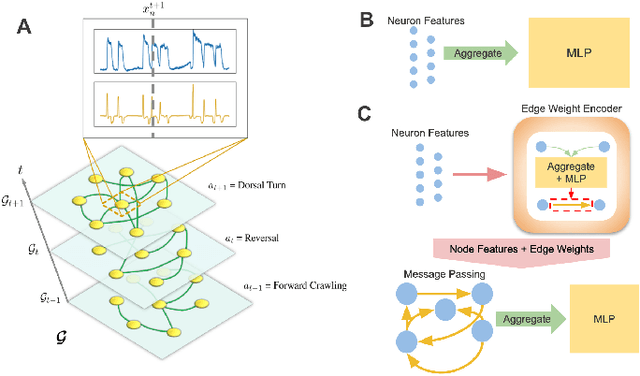
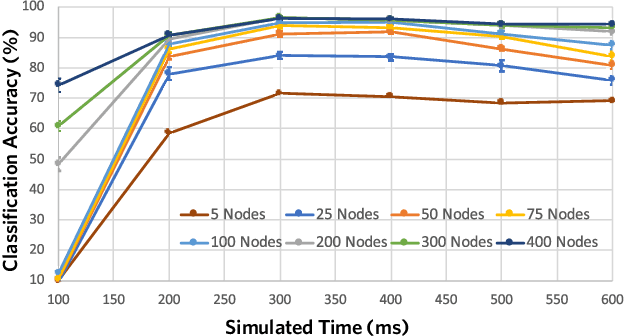
Abstract:Although a number of studies have explored deep learning in neuroscience, the application of these algorithms to neural systems on a microscopic scale, i.e. parameters relevant to lower scales of organization, remains relatively novel. Motivated by advances in whole-brain imaging, we examined the performance of deep learning models on microscopic neural dynamics and resulting emergent behaviors using calcium imaging data from the nematode C. elegans. We show that neural networks perform remarkably well on both neuron-level dynamics prediction, and behavioral state classification. In addition, we compared the performance of structure agnostic neural networks and graph neural networks to investigate if graph structure can be exploited as a favorable inductive bias. To perform this experiment, we designed a graph neural network which explicitly infers relations between neurons from neural activity and leverages the inferred graph structure during computations. In our experiments, we found that graph neural networks generally outperformed structure agnostic models and excel in generalization on unseen organisms, implying a potential path to generalizable machine learning in neuroscience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge