Umut Oztok
On Compiling DNNFs without Determinism
Sep 20, 2017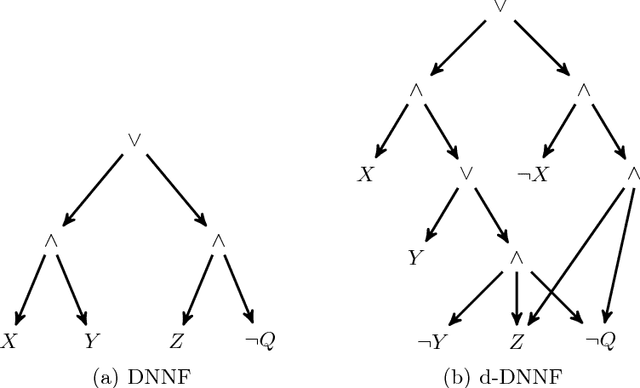
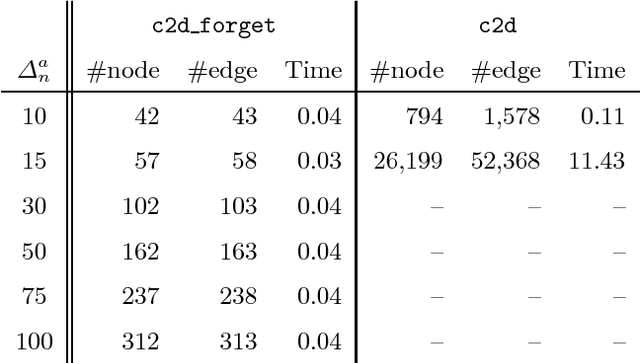
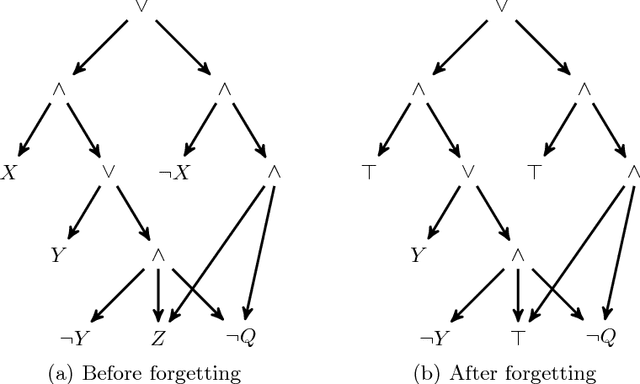
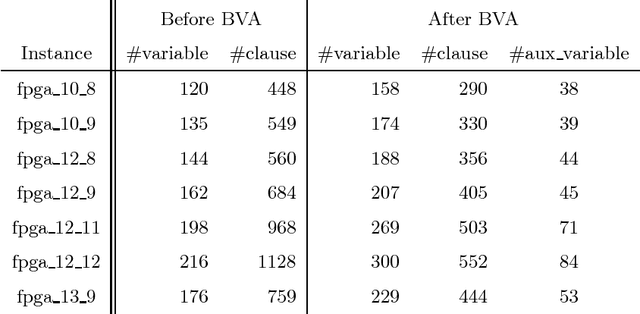
Abstract:State-of-the-art knowledge compilers generate deterministic subsets of DNNF, which have been recently shown to be exponentially less succinct than DNNF. In this paper, we propose a new method to compile DNNFs without enforcing determinism necessarily. Our approach is based on compiling deterministic DNNFs with the addition of auxiliary variables to the input formula. These variables are then existentially quantified from the deterministic structure in linear time, which would lead to a DNNF that is equivalent to the input formula and not necessarily deterministic. On the theoretical side, we show that the new method could generate exponentially smaller DNNFs than deterministic ones, even by adding a single auxiliary variable. Further, we show that various existing techniques that introduce auxiliary variables to the input formulas can be employed in our framework. On the practical side, we empirically demonstrate that our new method can significantly advance DNNF compilation on certain benchmarks.
Generating Explanations for Biomedical Queries
Sep 24, 2013
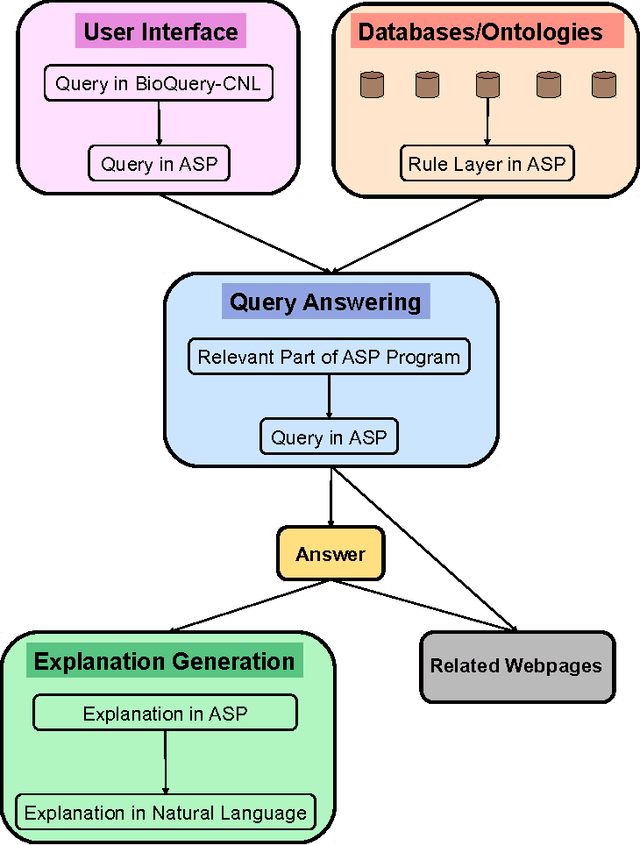
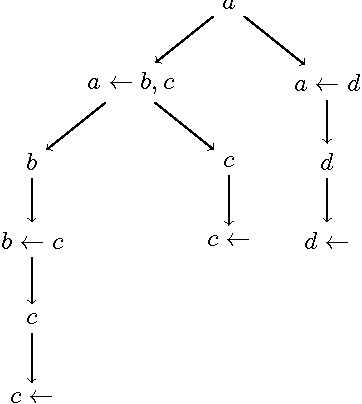
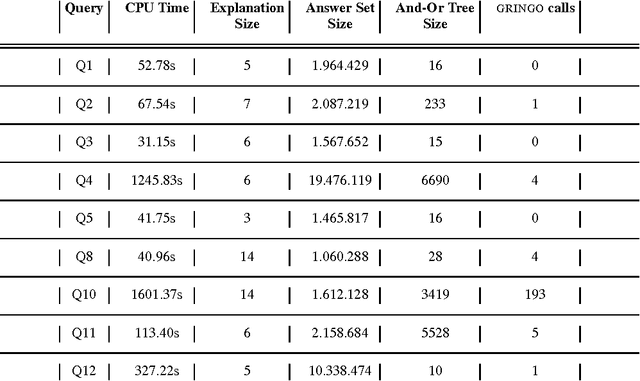
Abstract:We introduce novel mathematical models and algorithms to generate (shortest or k different) explanations for biomedical queries, using answer set programming. We implement these algorithms and integrate them in BIOQUERY-ASP. We illustrate the usefulness of these methods with some complex biomedical queries related to drug discovery, over the biomedical knowledge resources PHARMGKB, DRUGBANK, BIOGRID, CTD, SIDER, DISEASE ONTOLOGY and ORPHADATA. To appear in Theory and Practice of Logic Programming (TPLP).
Querying Biomedical Ontologies in Natural Language using Answer Set
Dec 09, 2010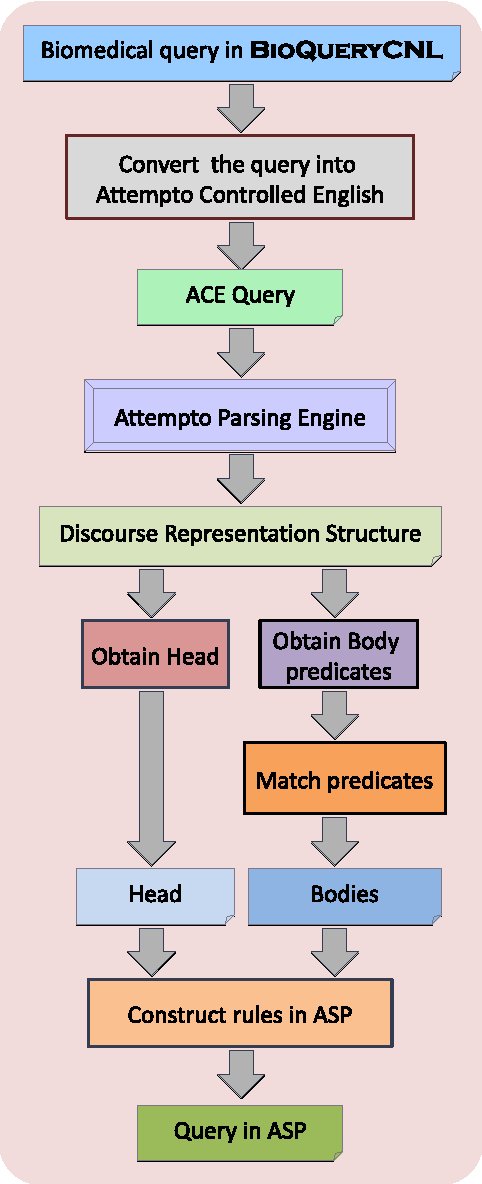
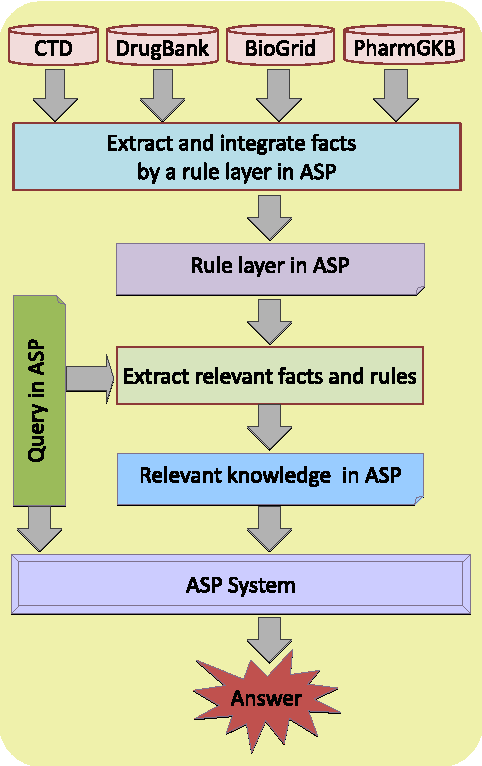
Abstract:In this work, we develop an intelligent user interface that allows users to enter biomedical queries in a natural language, and that presents the answers (possibly with explanations if requested) in a natural language. We develop a rule layer over biomedical ontologies and databases, and use automated reasoners to answer queries considering relevant parts of the rule layer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge