Uday Bondhugula
MLIR: A Compiler Infrastructure for the End of Moore's Law
Mar 01, 2020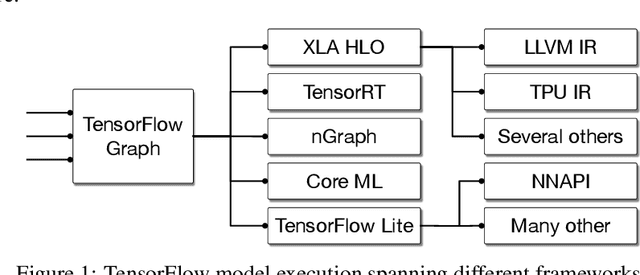
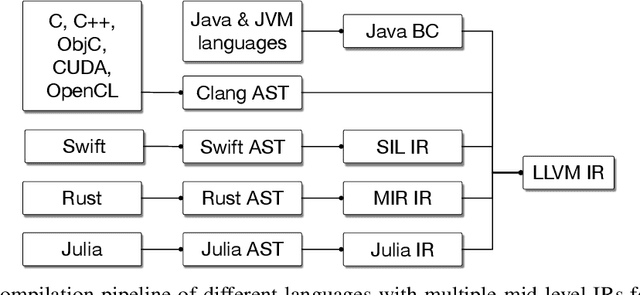
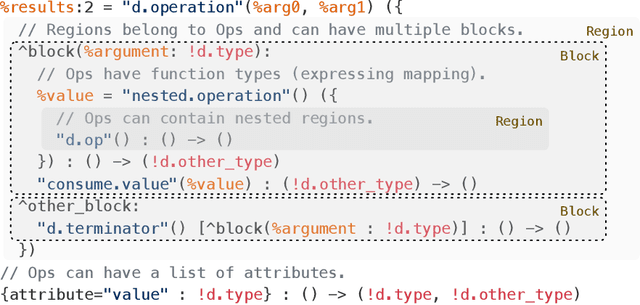
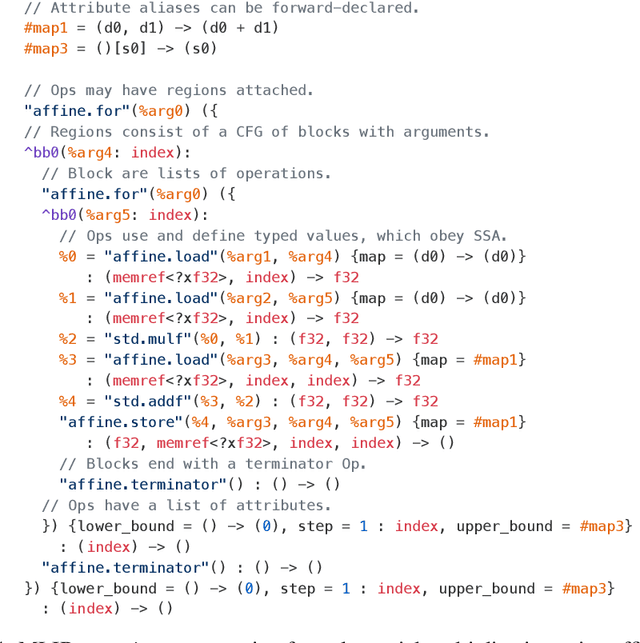
Abstract:This work presents MLIR, a novel approach to building reusable and extensible compiler infrastructure. MLIR aims to address software fragmentation, improve compilation for heterogeneous hardware, significantly reduce the cost of building domain specific compilers, and aid in connecting existing compilers together. MLIR facilitates the design and implementation of code generators, translators and optimizers at different levels of abstraction and also across application domains, hardware targets and execution environments. The contribution of this work includes (1) discussion of MLIR as a research artifact, built for extension and evolution, and identifying the challenges and opportunities posed by this novel design point in design, semantics, optimization specification, system, and engineering. (2) evaluation of MLIR as a generalized infrastructure that reduces the cost of building compilers-describing diverse use-cases to show research and educational opportunities for future programming languages, compilers, execution environments, and computer architecture. The paper also presents the rationale for MLIR, its original design principles, structures and semantics.
Optimizing the Linear Fascicle Evaluation Algorithm for Multi-Core and Many-Core Systems
May 14, 2019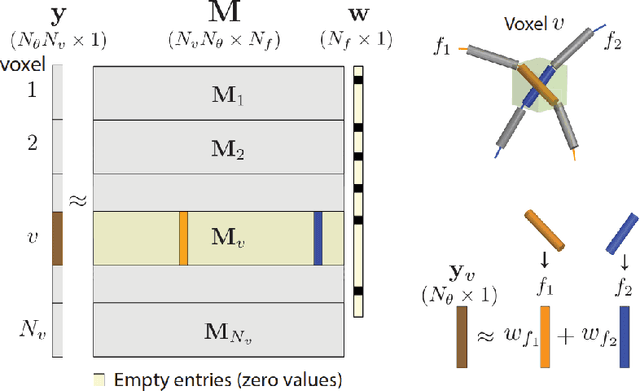
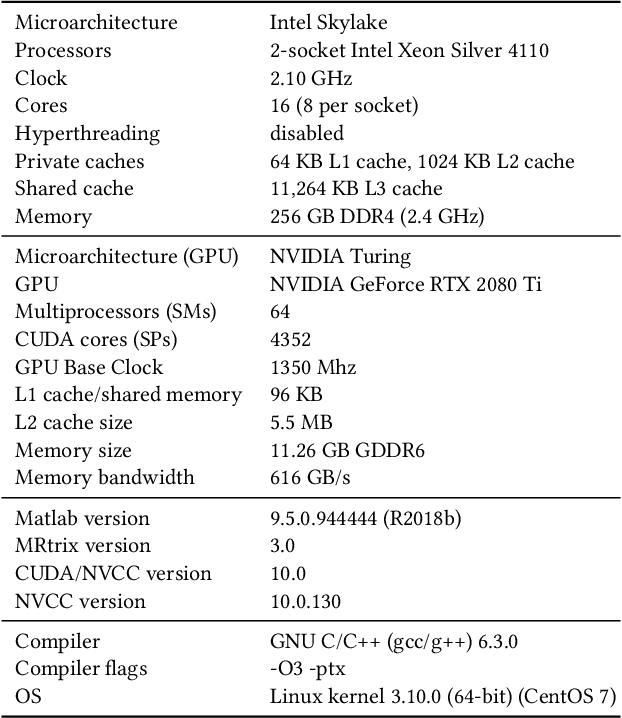
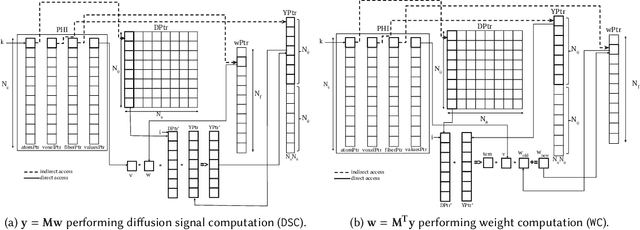
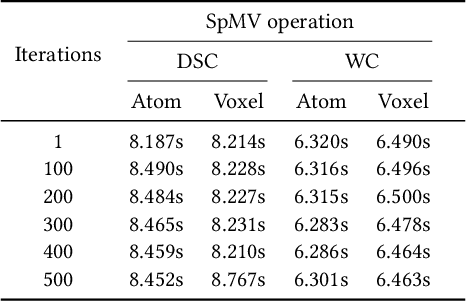
Abstract:Sparse matrix-vector multiplication (SpMV) operations are commonly used in various scientific applications. The performance of the SpMV operation often depends on exploiting regularity patterns in the matrix. Various representations have been proposed to minimize the memory bandwidth bottleneck arising from the irregular memory access pattern involved. Among recent representation techniques, tensor decomposition is a popular one used for very large but sparse matrices. Post sparse-tensor decomposition, the new representation involves indirect accesses, making it challenging to optimize for multi-cores and GPUs. Computational neuroscience algorithms often involve sparse datasets while still performing long-running computations on them. The LiFE application is a popular neuroscience algorithm used for pruning brain connectivity graphs. The datasets employed herein involve the Sparse Tucker Decomposition (STD), a widely used tensor decomposition method. Using this decomposition leads to irregular array references, making it very difficult to optimize for both CPUs and GPUs. Recent codes of the LiFE algorithm show that its SpMV operations are the key bottleneck for performance and scaling. In this work, we first propose target-independent optimizations to optimize these SpMV operations, followed by target-dependent optimizations for CPU and GPU systems. The target-independent techniques include: (1) standard compiler optimizations, (2) data restructuring methods, and (3) methods to partition computations among threads. Then we present the optimizations for CPUs and GPUs to exploit platform-specific speed. Our highly optimized CPU code obtain a speedup of 27.12x over the original sequential CPU code running on 16-core Intel Xeon (Skylake-based) system, and our optimized GPU code achieves a speedup of 5.2x over a reference optimized GPU code version on NVIDIA's GeForce RTX 2080 Ti GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge