Tomasz Szczepański
GEPAR3D: Geometry Prior-Assisted Learning for 3D Tooth Segmentation
Jul 31, 2025Abstract:Tooth segmentation in Cone-Beam Computed Tomography (CBCT) remains challenging, especially for fine structures like root apices, which is critical for assessing root resorption in orthodontics. We introduce GEPAR3D, a novel approach that unifies instance detection and multi-class segmentation into a single step tailored to improve root segmentation. Our method integrates a Statistical Shape Model of dentition as a geometric prior, capturing anatomical context and morphological consistency without enforcing restrictive adjacency constraints. We leverage a deep watershed method, modeling each tooth as a continuous 3D energy basin encoding voxel distances to boundaries. This instance-aware representation ensures accurate segmentation of narrow, complex root apices. Trained on publicly available CBCT scans from a single center, our method is evaluated on external test sets from two in-house and two public medical centers. GEPAR3D achieves the highest overall segmentation performance, averaging a Dice Similarity Coefficient (DSC) of 95.0% (+2.8% over the second-best method) and increasing recall to 95.2% (+9.5%) across all test sets. Qualitative analyses demonstrated substantial improvements in root segmentation quality, indicating significant potential for more accurate root resorption assessment and enhanced clinical decision-making in orthodontics. We provide the implementation and dataset at https://github.com/tomek1911/GEPAR3D.
Let Me DeCode You: Decoder Conditioning with Tabular Data
Jul 12, 2024



Abstract:Training deep neural networks for 3D segmentation tasks can be challenging, often requiring efficient and effective strategies to improve model performance. In this study, we introduce a novel approach, DeCode, that utilizes label-derived features for model conditioning to support the decoder in the reconstruction process dynamically, aiming to enhance the efficiency of the training process. DeCode focuses on improving 3D segmentation performance through the incorporation of conditioning embedding with learned numerical representation of 3D-label shape features. Specifically, we develop an approach, where conditioning is applied during the training phase to guide the network toward robust segmentation. When labels are not available during inference, our model infers the necessary conditioning embedding directly from the input data, thanks to a feed-forward network learned during the training phase. This approach is tested using synthetic data and cone-beam computed tomography (CBCT) images of teeth. For CBCT, three datasets are used: one publicly available and two in-house. Our results show that DeCode significantly outperforms traditional, unconditioned models in terms of generalization to unseen data, achieving higher accuracy at a reduced computational cost. This work represents the first of its kind to explore conditioning strategies in 3D data segmentation, offering a novel and more efficient method for leveraging annotated data. Our code, pre-trained models are publicly available at https://github.com/SanoScience/DeCode .
POTHER: Patch-Voted Deep Learning-based Chest X-ray Bias Analysis for COVID-19 Detection
Jan 23, 2022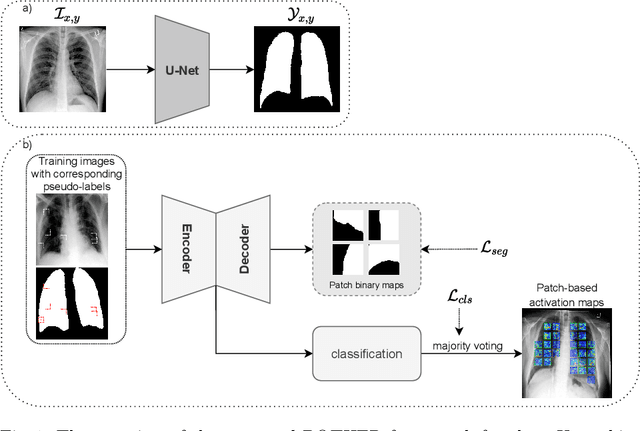
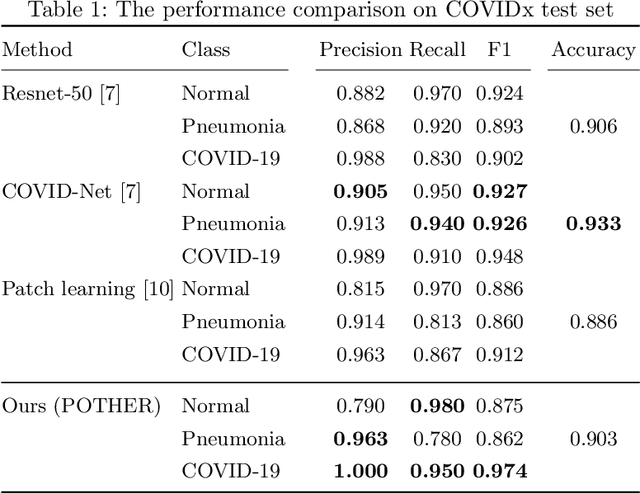
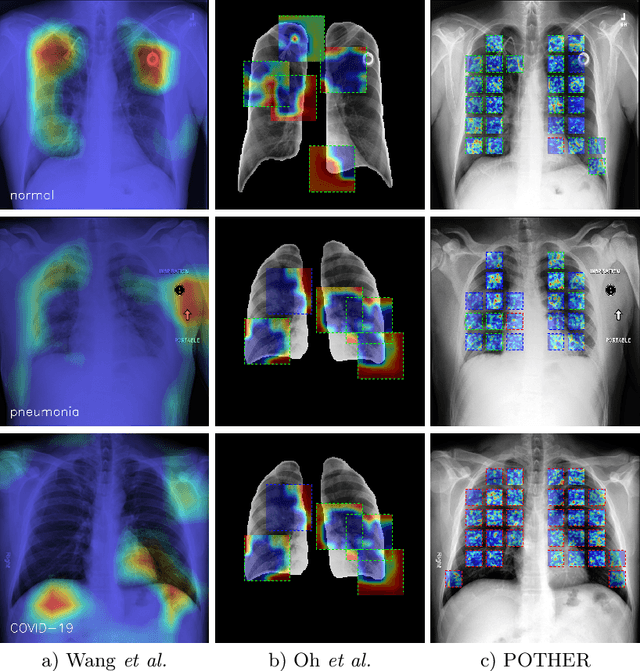
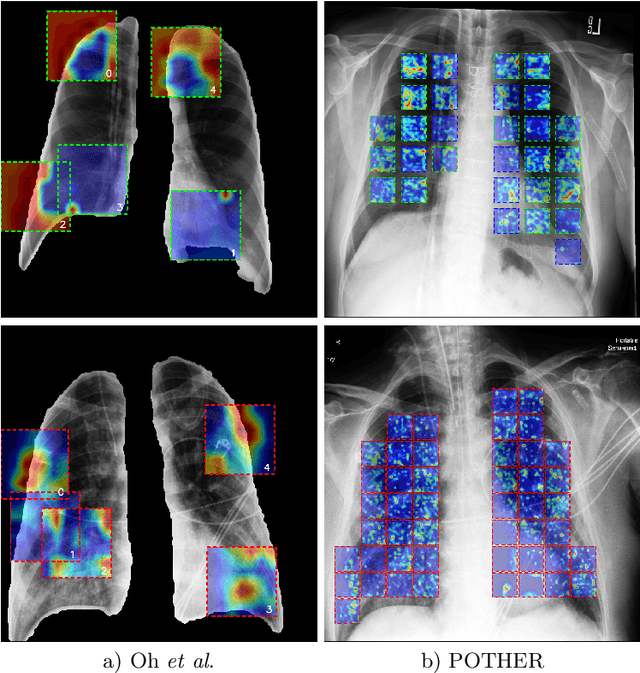
Abstract:A critical step in the fight against COVID-19, which continues to have a catastrophic impact on peoples lives, is the effective screening of patients presented in the clinics with severe COVID-19 symptoms. Chest radiography is one of the promising screening approaches. Many studies reported detecting COVID-19 in chest X-rays accurately using deep learning. A serious limitation of many published approaches is insufficient attention paid to explaining decisions made by deep learning models. Using explainable artificial intelligence methods, we demonstrate that model decisions may rely on confounding factors rather than medical pathology. After an analysis of potential confounding factors found on chest X-ray images, we propose a novel method to minimise their negative impact. We show that our proposed method is more robust than previous attempts to counter confounding factors such as ECG leads in chest X-rays that often influence model classification decisions. In addition to being robust, our method achieves results comparable to the state-of-the-art. The source code and pre-trained weights are publicly available (https://github.com/tomek1911/POTHER).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge