Tom George Grigg
Active Learning on Synthons for Molecular Design
May 19, 2025Abstract:Exhaustive virtual screening is highly informative but often intractable against the expensive objective functions involved in modern drug discovery. This problem is exacerbated in combinatorial contexts such as multi-vector expansion, where molecular spaces can quickly become ultra-large. Here, we introduce Scalable Active Learning via Synthon Acquisition (SALSA): a simple algorithm applicable to multi-vector expansion which extends pool-based active learning to non-enumerable spaces by factoring modeling and acquisition over synthon or fragment choices. Through experiments on ligand- and structure-based objectives, we highlight SALSA's sample efficiency, and its ability to scale to spaces of trillions of compounds. Further, we demonstrate application toward multi-parameter objective design tasks on three protein targets - finding SALSA-generated molecules have comparable chemical property profiles to known bioactives, and exhibit greater diversity and higher scores over an industry-leading generative approach.
Do Self-Supervised and Supervised Methods Learn Similar Visual Representations?
Oct 01, 2021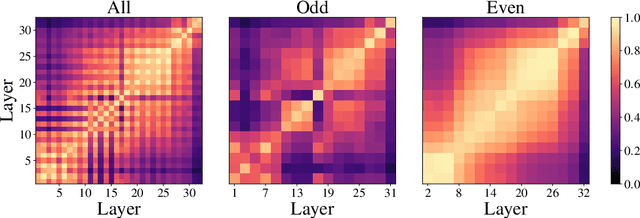
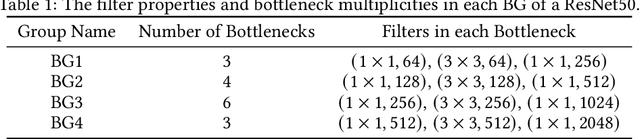
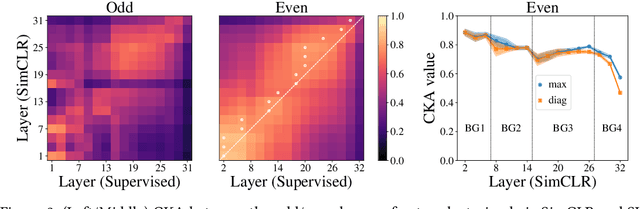
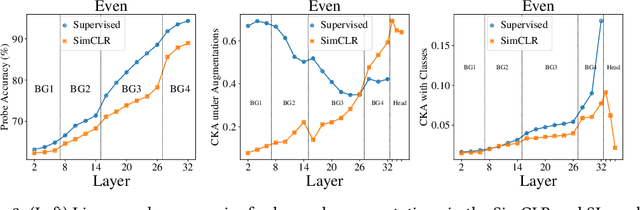
Abstract:Despite the success of a number of recent techniques for visual self-supervised deep learning, there remains limited investigation into the representations that are ultimately learned. By using recent advances in comparing neural representations, we explore in this direction by comparing a constrastive self-supervised algorithm (SimCLR) to supervision for simple image data in a common architecture. We find that the methods learn similar intermediate representations through dissimilar means, and that the representations diverge rapidly in the final few layers. We investigate this divergence, finding that it is caused by these layers strongly fitting to the distinct learning objectives. We also find that SimCLR's objective implicitly fits the supervised objective in intermediate layers, but that the reverse is not true. Our work particularly highlights the importance of the learned intermediate representations, and raises important questions for auxiliary task design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge