Tiffany M. Tang
Consensus dimension reduction via multi-view learning
Dec 16, 2025Abstract:A plethora of dimension reduction methods have been developed to visualize high-dimensional data in low dimensions. However, different dimension reduction methods often output different and possibly conflicting visualizations of the same data. This problem is further exacerbated by the choice of hyperparameters, which may substantially impact the resulting visualization. To obtain a more robust and trustworthy dimension reduction output, we advocate for a consensus approach, which summarizes multiple visualizations into a single consensus dimension reduction visualization. Here, we leverage ideas from multi-view learning in order to identify the patterns that are most stable or shared across the many different dimension reduction visualizations, or views, and subsequently visualize this shared structure in a single low-dimensional plot. We demonstrate that this consensus visualization effectively identifies and preserves the shared low-dimensional data structure through both simulated and real-world case studies. We further highlight our method's robustness to the choice of dimension reduction method and hyperparameters -- a highly-desirable property when working towards trustworthy and reproducible data science.
Interpretable Network-assisted Random Forest+
Sep 19, 2025Abstract:Machine learning algorithms often assume that training samples are independent. When data points are connected by a network, the induced dependency between samples is both a challenge, reducing effective sample size, and an opportunity to improve prediction by leveraging information from network neighbors. Multiple methods taking advantage of this opportunity are now available, but many, including graph neural networks, are not easily interpretable, limiting their usefulness for understanding how a model makes its predictions. Others, such as network-assisted linear regression, are interpretable but often yield substantially worse prediction performance. We bridge this gap by proposing a family of flexible network-assisted models built upon a generalization of random forests (RF+), which achieves highly-competitive prediction accuracy and can be interpreted through feature importance measures. In particular, we develop a suite of interpretation tools that enable practitioners to not only identify important features that drive model predictions, but also quantify the importance of the network contribution to prediction. Importantly, we provide both global and local importance measures as well as sample influence measures to assess the impact of a given observation. This suite of tools broadens the scope and applicability of network-assisted machine learning for high-impact problems where interpretability and transparency are essential.
Local MDI+: Local Feature Importances for Tree-Based Models
Jun 10, 2025Abstract:Tree-based ensembles such as random forests remain the go-to for tabular data over deep learning models due to their prediction performance and computational efficiency. These advantages have led to their widespread deployment in high-stakes domains, where interpretability is essential for ensuring trustworthy predictions. This has motivated the development of popular local (i.e. sample-specific) feature importance (LFI) methods such as LIME and TreeSHAP. However, these approaches rely on approximations that ignore the model's internal structure and instead depend on potentially unstable perturbations. These issues are addressed in the global setting by MDI+, a feature importance method which exploits an equivalence between decision trees and linear models on a transformed node basis. However, the global MDI+ scores are not able to explain predictions when faced with heterogeneous individual characteristics. To address this gap, we propose Local MDI+ (LMDI+), a novel extension of the MDI+ framework to the sample specific setting. LMDI+ outperforms existing baselines LIME and TreeSHAP in identifying instance-specific signal features, averaging a 10% improvement in downstream task performance across twelve real-world benchmark datasets. It further demonstrates greater stability by consistently producing similar instance-level feature importance rankings across multiple random forest fits. Finally, LMDI+ enables local interpretability use cases, including the identification of closer counterfactuals and the discovery of homogeneous subgroups.
Unsupervised Machine Learning for Scientific Discovery: Workflow and Best Practices
Jun 05, 2025



Abstract:Unsupervised machine learning is widely used to mine large, unlabeled datasets to make data-driven discoveries in critical domains such as climate science, biomedicine, astronomy, chemistry, and more. However, despite its widespread utilization, there is a lack of standardization in unsupervised learning workflows for making reliable and reproducible scientific discoveries. In this paper, we present a structured workflow for using unsupervised learning techniques in science. We highlight and discuss best practices starting with formulating validatable scientific questions, conducting robust data preparation and exploration, using a range of modeling techniques, performing rigorous validation by evaluating the stability and generalizability of unsupervised learning conclusions, and promoting effective communication and documentation of results to ensure reproducible scientific discoveries. To illustrate our proposed workflow, we present a case study from astronomy, seeking to refine globular clusters of Milky Way stars based upon their chemical composition. Our case study highlights the importance of validation and illustrates how the benefits of a carefully-designed workflow for unsupervised learning can advance scientific discovery.
MDI+: A Flexible Random Forest-Based Feature Importance Framework
Jul 04, 2023



Abstract:Mean decrease in impurity (MDI) is a popular feature importance measure for random forests (RFs). We show that the MDI for a feature $X_k$ in each tree in an RF is equivalent to the unnormalized $R^2$ value in a linear regression of the response on the collection of decision stumps that split on $X_k$. We use this interpretation to propose a flexible feature importance framework called MDI+. Specifically, MDI+ generalizes MDI by allowing the analyst to replace the linear regression model and $R^2$ metric with regularized generalized linear models (GLMs) and metrics better suited for the given data structure. Moreover, MDI+ incorporates additional features to mitigate known biases of decision trees against additive or smooth models. We further provide guidance on how practitioners can choose an appropriate GLM and metric based upon the Predictability, Computability, Stability framework for veridical data science. Extensive data-inspired simulations show that MDI+ significantly outperforms popular feature importance measures in identifying signal features. We also apply MDI+ to two real-world case studies on drug response prediction and breast cancer subtype classification. We show that MDI+ extracts well-established predictive genes with significantly greater stability compared to existing feature importance measures. All code and models are released in a full-fledged python package on Github.
Feature Selection for Data Integration with Mixed Multi-view Data
Mar 27, 2019
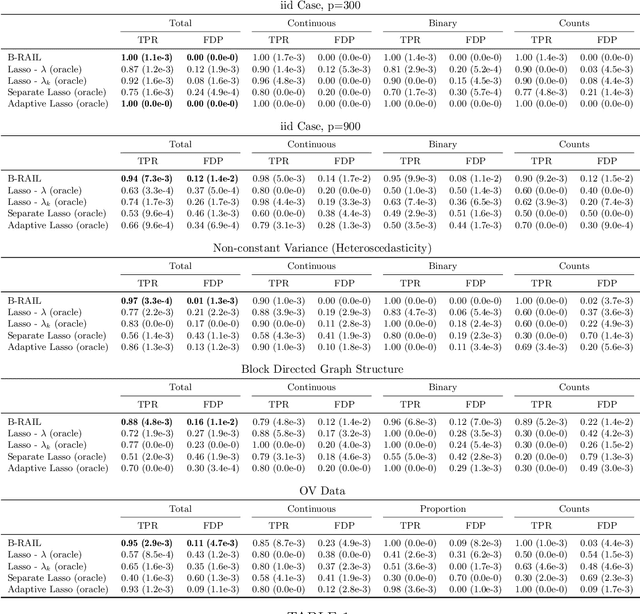
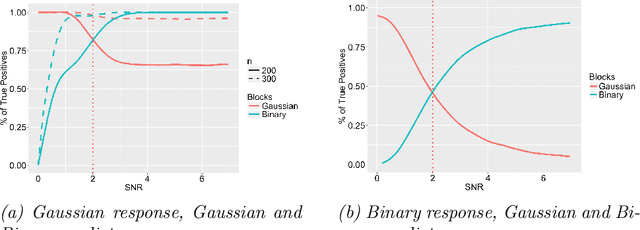
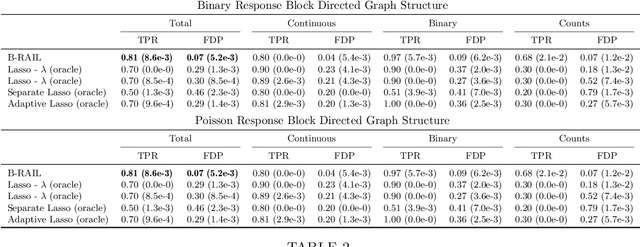
Abstract:Data integration methods that analyze multiple sources of data simultaneously can often provide more holistic insights than can separate inquiries of each data source. Motivated by the advantages of data integration in the era of "big data", we investigate feature selection for high-dimensional multi-view data with mixed data types (e.g. continuous, binary, count-valued). This heterogeneity of multi-view data poses numerous challenges for existing feature selection methods. However, after critically examining these issues through empirical and theoretically-guided lenses, we develop a practical solution, the Block Randomized Adaptive Iterative Lasso (B-RAIL), which combines the strengths of the randomized Lasso, adaptive weighting schemes, and stability selection. B-RAIL serves as a versatile data integration method for sparse regression and graph selection, and we demonstrate the effectiveness of B-RAIL through extensive simulations and a case study to infer the ovarian cancer gene regulatory network. In this case study, B-RAIL successfully identifies well-known biomarkers associated with ovarian cancer and hints at novel candidates for future ovarian cancer research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge