Tianwei Yu
Learning, Solving and Optimizing PDEs with TensorGalerkin: an efficient high-performance Galerkin assembly algorithm
Feb 04, 2026Abstract:We present a unified algorithmic framework for the numerical solution, constrained optimization, and physics-informed learning of PDEs with a variational structure. Our framework is based on a Galerkin discretization of the underlying variational forms, and its high efficiency stems from a novel highly-optimized and GPU-compliant TensorGalerkin framework for linear system assembly (stiffness matrices and load vectors). TensorGalerkin operates by tensorizing element-wise operations within a Python-level Map stage and then performs global reduction with a sparse matrix multiplication that performs message passing on the mesh-induced sparsity graph. It can be seamlessly employed downstream as i) a highly-efficient numerical PDEs solver, ii) an end-to-end differentiable framework for PDE-constrained optimization, and iii) a physics-informed operator learning algorithm for PDEs. With multiple benchmarks, including 2D and 3D elliptic, parabolic, and hyperbolic PDEs on unstructured meshes, we demonstrate that the proposed framework provides significant computational efficiency and accuracy gains over a variety of baselines in all the targeted downstream applications.
Fast Gibbs Sampling on Bayesian Hidden Markov Model with Missing Observations
Jan 04, 2026Abstract:The Hidden Markov Model (HMM) is a widely-used statistical model for handling sequential data. However, the presence of missing observations in real-world datasets often complicates the application of the model. The EM algorithm and Gibbs samplers can be used to estimate the model, yet suffering from various problems including non-convexity, high computational complexity and slow mixing. In this paper, we propose a collapsed Gibbs sampler that efficiently samples from HMMs' posterior by integrating out both the missing observations and the corresponding latent states. The proposed sampler is fast due to its three advantages. First, it achieves an estimation accuracy that is comparable to existing methods. Second, it can produce a larger Effective Sample Size (ESS) per iteration, which can be justified theoretically and numerically. Third, when the number of missing entries is large, the sampler has a significant smaller computational complexity per iteration compared to other methods, thus is faster computationally. In summary, the proposed sampling algorithm is fast both computationally and theoretically and is particularly advantageous when there are a lot of missing entries. Finally, empirical evaluations based on numerical simulations and real data analysis demonstrate that the proposed algorithm consistently outperforms existing algorithms in terms of time complexity and sampling efficiency (measured in ESS).
Learning data representation using modified autoencoder for the integrative analysis of multi-omics data
Jun 18, 2019
Abstract:In integrative analyses of omics data, it is often of interest to extract data embedding from one data type that best reflect relations with another data type. This task is traditionally fulfilled by linear methods such as canonical correlation and partial least squares. However, information contained in one data type pertaining to the other data type may not be in the linear form. Deep learning provides a convenient alternative to extract nonlinear information. Here we develop a method Autoencoder-based Integrative Multi-omics data Embedding (AIME) to extract such information. Using a real gene expression - methylation dataset, we show that AIME extracted meaningful information that the linear approach could not find. The R implementation is available at http://web1.sph.emory.edu/users/tyu8/AIME/.
forgeNet: A graph deep neural network model using tree-based ensemble classifiers for feature extraction
May 23, 2019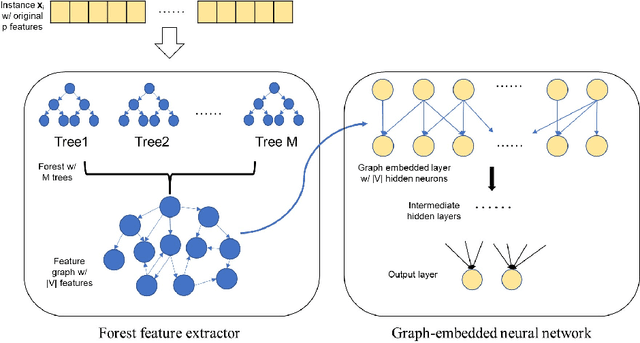

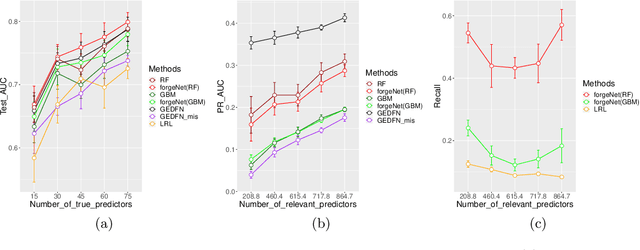

Abstract:A unique challenge in predictive model building for omics data has been the small number of samples $(n)$ versus the large amount of features $(p)$. This "$n\ll p$" property brings difficulties for disease outcome classification using deep learning techniques. Sparse learning by incorporating external gene network information such as the graph-embedded deep feedforward network (GEDFN) model has been a solution to this issue. However, such methods require an existing feature graph, and potential mis-specification of the feature graph can be harmful on classification and feature selection. To address this limitation and develop a robust classification model without relying on external knowledge, we propose a \underline{for}est \underline{g}raph-\underline{e}mbedded deep feedforward \underline{net}work (forgeNet) model, to integrate the GEDFN architecture with a forest feature graph extractor, so that the feature graph can be learned in a supervised manner and specifically constructed for a given prediction task. To validate the method's capability, we experimented the forgeNet model with both synthetic and real datasets. The resulting high classification accuracy suggests that the method is a valuable addition to sparse deep learning models for omics data.
Nonlinear variable selection with continuous outcome: a nonparametric incremental forward stagewise approach
May 26, 2018



Abstract:We present a method of variable selection for the sparse generalized additive model. The method doesn't assume any specific functional form, and can select from a large number of candidates. It takes the form of incremental forward stagewise regression. Given no functional form is assumed, we devised an approach termed roughening to adjust the residuals in the iterations. In simulations, we show the new method is competitive against popular machine learning approaches. We also demonstrate its performance using some real datasets. The method is available as a part of the nlnet package on CRAN https://cran.r-project.org/package=nlnet.
A graph-embedded deep feedforward network for disease outcome classification and feature selection using gene expression data
Feb 12, 2018



Abstract:Gene expression data represents a unique challenge in predictive model building, because of the small number of samples $(n)$ compared to the huge amount of features $(p)$. This "$n<<p$" property has hampered application of deep learning techniques for disease outcome classification. Sparse learning by incorporating external gene network information could be a potential solution to this issue. Still, the problem is very challenging because (1) there are tens of thousands of features and only hundreds of training samples, (2) the scale-free structure of the gene network is unfriendly to the setup of convolutional neural networks. To address these issues and build a robust classification model, we propose the Graph-Embedded Deep Feedforward Networks (GEDFN), to integrate external relational information of features into the deep neural network architecture. The method is able to achieve sparse connection between network layers to prevent overfitting. To validate the method's capability, we conducted both simulation experiments and a real data analysis using a breast cancer RNA-seq dataset from The Cancer Genome Atlas (TCGA). The resulting high classification accuracy and easily interpretable feature selection results suggest the method is a useful addition to the current classification models and feature selection procedures. The method is available at https://github.com/yunchuankong/NetworkNeuralNetwork.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge