Sukrit Gupta
CASPER: Cross-modal Alignment of Spatial and single-cell Profiles for Expression Recovery
Nov 19, 2025



Abstract:Spatial Transcriptomics enables mapping of gene expression within its native tissue context, but current platforms measure only a limited set of genes due to experimental constraints and excessive costs. To overcome this, computational models integrate Single-Cell RNA Sequencing data with Spatial Transcriptomics to predict unmeasured genes. We propose CASPER, a cross-attention based framework that predicts unmeasured gene expression in Spatial Transcriptomics by leveraging centroid-level representations from Single-Cell RNA Sequencing. We performed rigorous testing over four state-of-the-art Spatial Transcriptomics/Single-Cell RNA Sequencing dataset pairs across four existing baseline models. CASPER shows significant improvement in nine out of the twelve metrics for our experiments. This work paves the way for further work in Spatial Transcriptomics to Single-Cell RNA Sequencing modality translation. The code for CASPER is available at https://github.com/AI4Med-Lab/CASPER.
WaveFuse-AL: Cyclical and Performance-Adaptive Multi-Strategy Active Learning for Medical Images
Nov 19, 2025Abstract:Active learning reduces annotation costs in medical imaging by strategically selecting the most informative samples for labeling. However, individual acquisition strategies often exhibit inconsistent behavior across different stages of the active learning cycle. We propose Cyclical and Performance-Adaptive Multi-Strategy Active Learning (WaveFuse-AL), a novel framework that adaptively fuses multiple established acquisition strategies-BALD, BADGE, Entropy, and CoreSet throughout the learning process. WaveFuse-AL integrates cyclical (sinusoidal) temporal priors with performance-driven adaptation to dynamically adjust strategy importance over time. We evaluate WaveFuse-AL on three medical imaging benchmarks: APTOS-2019 (multi-class classification), RSNA Pneumonia Detection (binary classification), and ISIC-2018 (skin lesion segmentation). Experimental results demonstrate that WaveFuse-AL consistently outperforms both single-strategy and alternating-strategy baselines, achieving statistically significant performance improvements (on ten out of twelve metric measurements) while maximizing the utility of limited annotation budgets.
Discovering robust biomarkers of neurological disorders from functional MRI using graph neural networks: A Review
May 01, 2024



Abstract:Graph neural networks (GNN) have emerged as a popular tool for modelling functional magnetic resonance imaging (fMRI) datasets. Many recent studies have reported significant improvements in disorder classification performance via more sophisticated GNN designs and highlighted salient features that could be potential biomarkers of the disorder. In this review, we provide an overview of how GNN and model explainability techniques have been applied on fMRI datasets for disorder prediction tasks, with a particular emphasis on the robustness of biomarkers produced for neurodegenerative diseases and neuropsychiatric disorders. We found that while most studies have performant models, salient features highlighted in these studies vary greatly across studies on the same disorder and little has been done to evaluate their robustness. To address these issues, we suggest establishing new standards that are based on objective evaluation metrics to determine the robustness of these potential biomarkers. We further highlight gaps in the existing literature and put together a prediction-attribution-evaluation framework that could set the foundations for future research on improving the robustness of potential biomarkers discovered via GNNs.
Deep ConvLSTM with self-attention for human activity decoding using wearables
May 02, 2020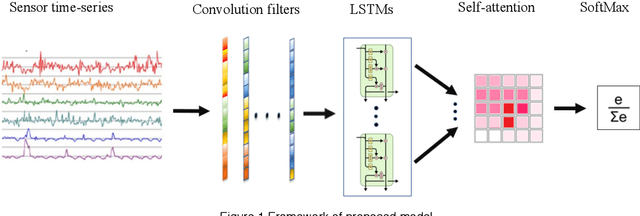
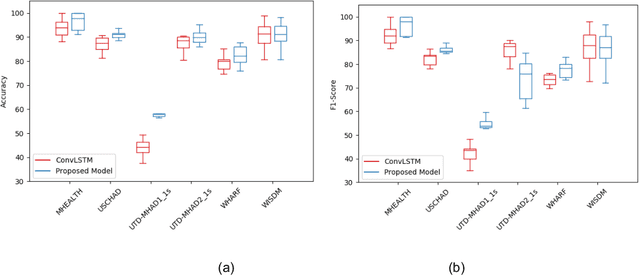

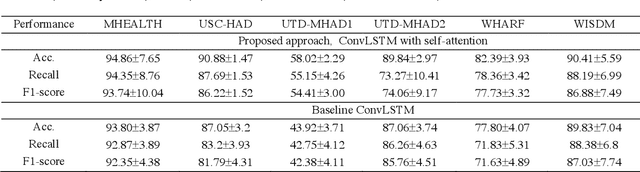
Abstract:Decoding human activity accurately from wearable sensors can aid in applications related to healthcare and context awareness. The present approaches in this domain use recurrent and/or convolutional models to capture the spatio-temporal features from time series data from multiple sensors. We propose a deep neural network architecture that not only captures the spatio-temporal features of multiple sensor time series data, but also selects, learns important time points by utilizing a self-attention mechanism. We show the validity of the proposed approach across different data sampling strategies on six public datasets and demonstrate that the self-attention mechanism gave significant improvement in performance over deep networks using a combination of recurrent and convolution networks. We also show that the proposed approach gave a statistically significant performance enhancement over previous state-of-the-art methods for the tested datasets. The proposed methods open avenues for better decoding of human activity from multiple body sensors over extended periods of time. The code implementation for the proposed model is available at https://github.com/isukrit/encodingHumanActivity
3D Deep Learning on Medical Images: A Review
Apr 01, 2020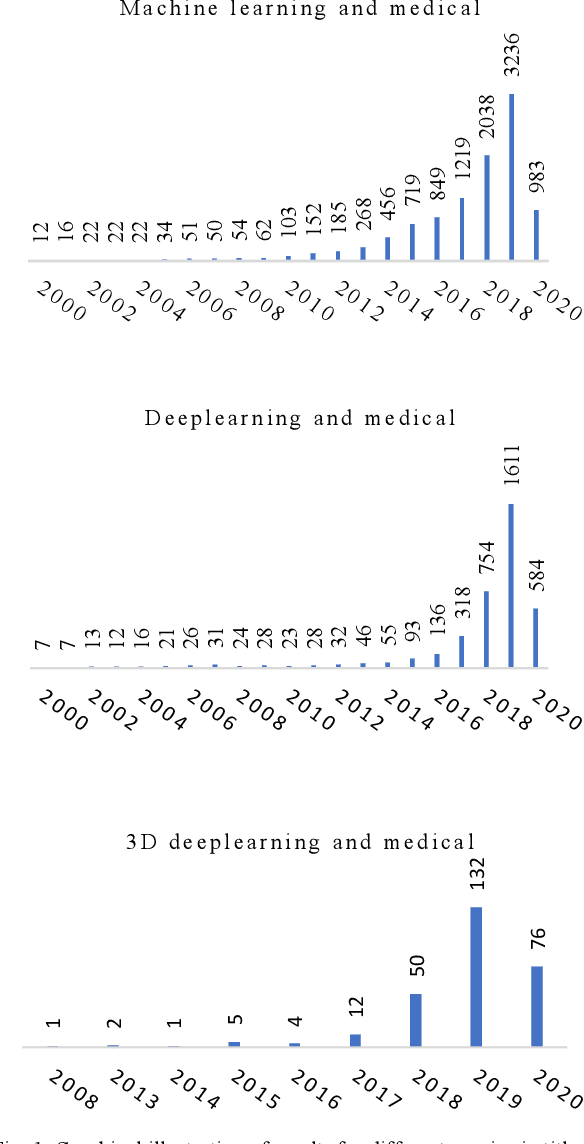
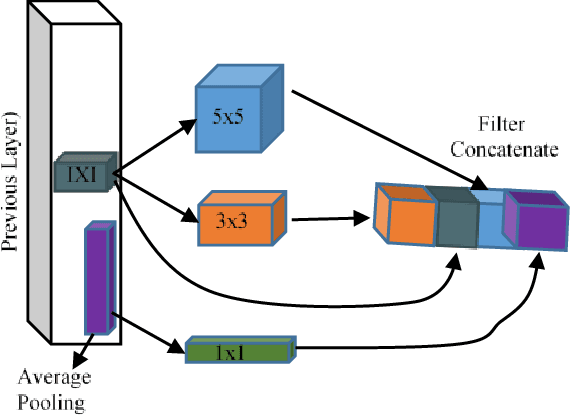
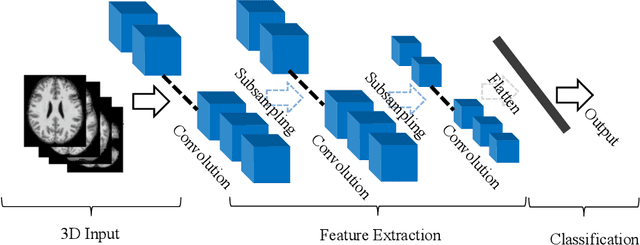
Abstract:The rapid advancements in machine learning, graphics processing technologies and availability of medical imaging data has led to a rapid increase in use of machine learning models in the medical domain. This was exacerbated by the rapid advancements in convolutional neural network (CNN) based architectures, which were adopted by the medical imaging community to assist clinicians in disease diagnosis. Since the grand success of AlexNet in 2012, CNNs have been increasingly used in medical image analysis to improve the efficiency of human clinicians. In recent years, three-dimensional (3D) CNNs have been employed for analysis of medical images. In this paper, we trace the history of how the 3D CNN was developed from its machine learning roots, brief mathematical description of 3D CNN and the preprocessing steps required for medical images before feeding them to 3D CNNs. We review the significant research in the field of 3D medical imaging analysis using 3D CNNs (and its variants) in different medical areas such as classification, segmentation, detection, and localization. We conclude by discussing the challenges associated with the use of 3D CNNs in the medical imaging domain (and the use of deep learning models, in general) and possible future trends in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge