Subrota Kumar Mondal
Batch Augmentation with Unimodal Fine-tuning for Multimodal Learning
May 10, 2025Abstract:This paper proposes batch augmentation with unimodal fine-tuning to detect the fetus's organs from ultrasound images and associated clinical textual information. We also prescribe pre-training initial layers with investigated medical data before the multimodal training. At first, we apply a transferred initialization with the unimodal image portion of the dataset with batch augmentation. This step adjusts the initial layer weights for medical data. Then, we apply neural networks (NNs) with fine-tuned initial layers to images in batches with batch augmentation to obtain features. We also extract information from descriptions of images. We combine this information with features obtained from images to train the head layer. We write a dataloader script to load the multimodal data and use existing unimodal image augmentation techniques with batch augmentation for the multimodal data. The dataloader brings a new random augmentation for each batch to get a good generalization. We investigate the FPU23 ultrasound and UPMC Food-101 multimodal datasets. The multimodal large language model (LLM) with the proposed training provides the best results among the investigated methods. We receive near state-of-the-art (SOTA) performance on the UPMC Food-101 dataset. We share the scripts of the proposed method with traditional counterparts at the following repository: github.com/dipuk0506/multimodal
Uncertainty Aware Neural Network from Similarity and Sensitivity
Apr 27, 2023Abstract:Researchers have proposed several approaches for neural network (NN) based uncertainty quantification (UQ). However, most of the approaches are developed considering strong assumptions. Uncertainty quantification algorithms often perform poorly in an input domain and the reason for poor performance remains unknown. Therefore, we present a neural network training method that considers similar samples with sensitivity awareness in this paper. In the proposed NN training method for UQ, first, we train a shallow NN for the point prediction. Then, we compute the absolute differences between prediction and targets and train another NN for predicting those absolute differences or absolute errors. Domains with high average absolute errors represent a high uncertainty. In the next step, we select each sample in the training set one by one and compute both prediction and error sensitivities. Then we select similar samples with sensitivity consideration and save indexes of similar samples. The ranges of an input parameter become narrower when the output is highly sensitive to that parameter. After that, we construct initial uncertainty bounds (UB) by considering the distribution of sensitivity aware similar samples. Prediction intervals (PIs) from initial uncertainty bounds are larger and cover more samples than required. Therefore, we train bound correction NN. As following all the steps for finding UB for each sample requires a lot of computation and memory access, we train a UB computation NN. The UB computation NN takes an input sample and provides an uncertainty bound. The UB computation NN is the final product of the proposed approach. Scripts of the proposed method are available in the following GitHub repository: github.com/dipuk0506/UQ
CoV-TI-Net: Transferred Initialization with Modified End Layer for COVID-19 Diagnosis
Sep 20, 2022
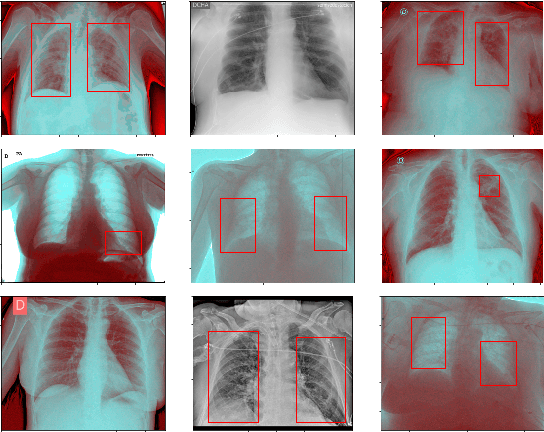
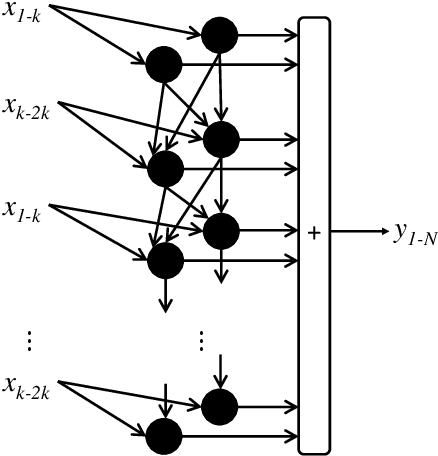
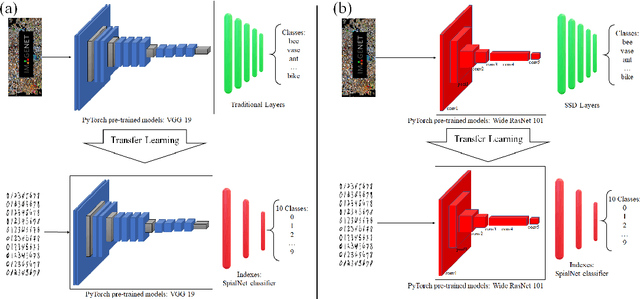
Abstract:This paper proposes transferred initialization with modified fully connected layers for COVID-19 diagnosis. Convolutional neural networks (CNN) achieved a remarkable result in image classification. However, training a high-performing model is a very complicated and time-consuming process because of the complexity of image recognition applications. On the other hand, transfer learning is a relatively new learning method that has been employed in many sectors to achieve good performance with fewer computations. In this research, the PyTorch pre-trained models (VGG19\_bn and WideResNet -101) are applied in the MNIST dataset for the first time as initialization and with modified fully connected layers. The employed PyTorch pre-trained models were previously trained in ImageNet. The proposed model is developed and verified in the Kaggle notebook, and it reached the outstanding accuracy of 99.77% without taking a huge computational time during the training process of the network. We also applied the same methodology to the SIIM-FISABIO-RSNA COVID-19 Detection dataset and achieved 80.01% accuracy. In contrast, the previous methods need a huge compactional time during the training process to reach a high-performing model. Codes are available at the following link: github.com/dipuk0506/SpinalNet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge