Stephen L. Rathbun
Multi-Task Adversarial Learning for Treatment Effect Estimation in Basket Trials
Mar 10, 2022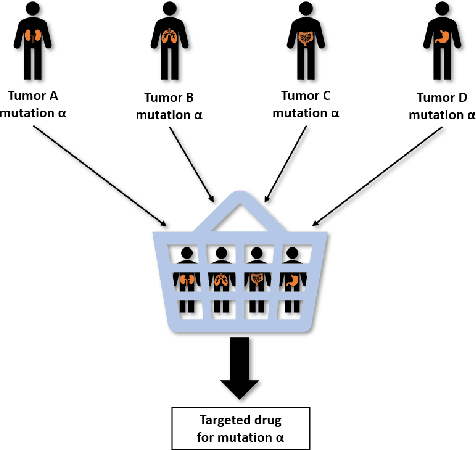
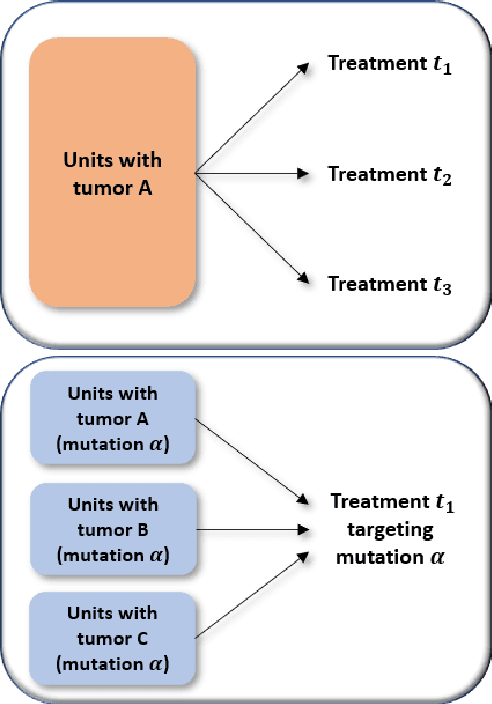
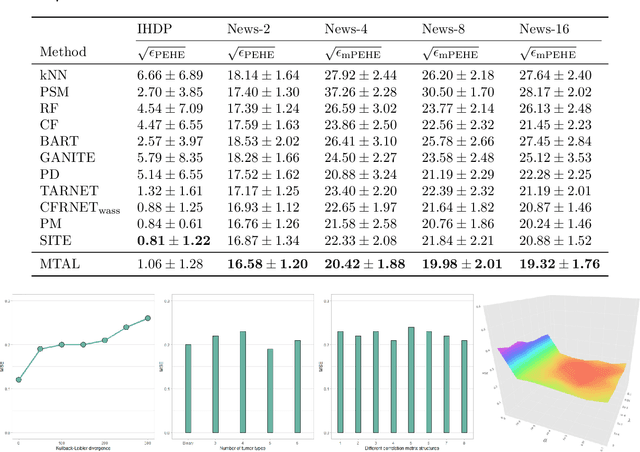
Abstract:Estimating treatment effects from observational data provides insights about causality guiding many real-world applications such as different clinical study designs, which are the formulations of trials, experiments, and observational studies in medical, clinical, and other types of research. In this paper, we describe causal inference for application in a novel clinical design called basket trial that tests how well a new drug works in patients who have different types of cancer that all have the same mutation. We propose a multi-task adversarial learning (MTAL) method, which incorporates feature selection multi-task representation learning and adversarial learning to estimate potential outcomes across different tumor types for patients sharing the same genetic mutation but having different tumor types. In our paper, the basket trial is employed as an intuitive example to present this new causal inference setting. This new causal inference setting includes, but is not limited to basket trials. This setting has the same challenges as the traditional causal inference problem, i.e., missing counterfactual outcomes under different subgroups and treatment selection bias due to confounders. We present the practical advantages of our MTAL method for the analysis of synthetic basket trial data and evaluate the proposed estimator on two benchmarks, IHDP and News. The results demonstrate the superiority of our MTAL method over the competing state-of-the-art methods.
Graph Infomax Adversarial Learning for Treatment Effect Estimation with Networked Observational Data
Jun 05, 2021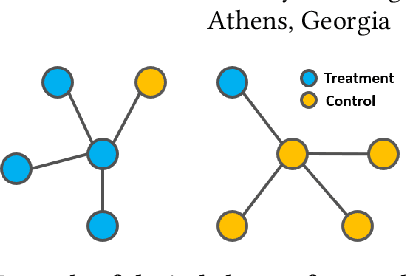
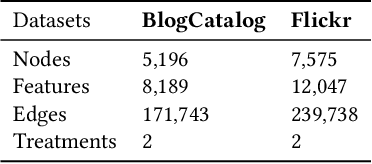
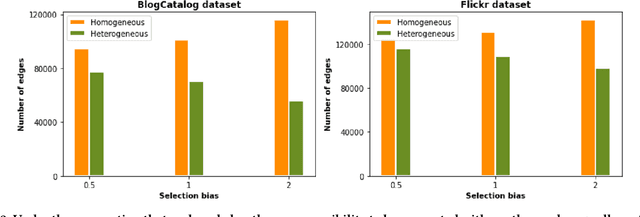
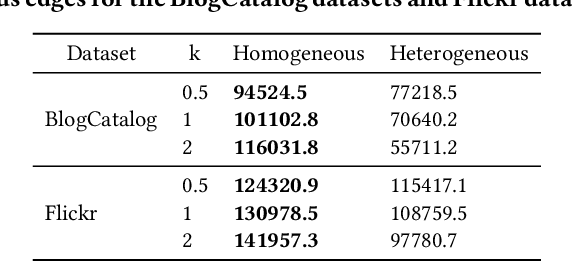
Abstract:Treatment effect estimation from observational data is a critical research topic across many domains. The foremost challenge in treatment effect estimation is how to capture hidden confounders. Recently, the growing availability of networked observational data offers a new opportunity to deal with the issue of hidden confounders. Unlike networked data in traditional graph learning tasks, such as node classification and link detection, the networked data under the causal inference problem has its particularity, i.e., imbalanced network structure. In this paper, we propose a Graph Infomax Adversarial Learning (GIAL) model for treatment effect estimation, which makes full use of the network structure to capture more information by recognizing the imbalance in network structure. We evaluate the performance of our GIAL model on two benchmark datasets, and the results demonstrate superiority over the state-of-the-art methods.
Matching in Selective and Balanced Representation Space for Treatment Effects Estimation
Sep 15, 2020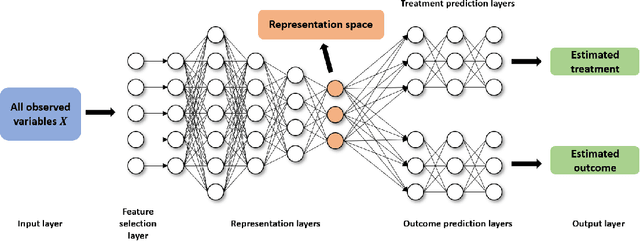
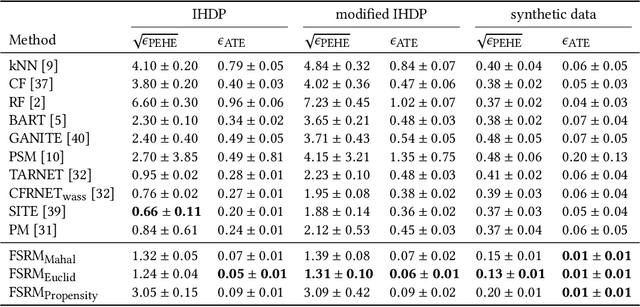
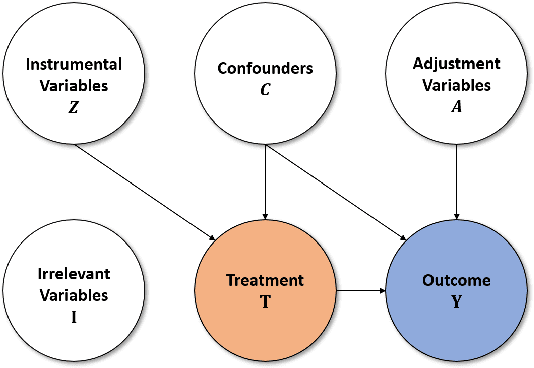
Abstract:The dramatically growing availability of observational data is being witnessed in various domains of science and technology, which facilitates the study of causal inference. However, estimating treatment effects from observational data is faced with two major challenges, missing counterfactual outcomes and treatment selection bias. Matching methods are among the most widely used and fundamental approaches to estimating treatment effects, but existing matching methods have poor performance when facing data with high dimensional and complicated variables. We propose a feature selection representation matching (FSRM) method based on deep representation learning and matching, which maps the original covariate space into a selective, nonlinear, and balanced representation space, and then conducts matching in the learned representation space. FSRM adopts deep feature selection to minimize the influence of irrelevant variables for estimating treatment effects and incorporates a regularizer based on the Wasserstein distance to learn balanced representations. We evaluate the performance of our FSRM method on three datasets, and the results demonstrate superiority over the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge