Sol Lee
Diagnose Like A REAL Pathologist: An Uncertainty-Focused Approach for Trustworthy Multi-Resolution Multiple Instance Learning
Nov 09, 2025Abstract:With the increasing demand for histopathological specimen examination and diagnostic reporting, Multiple Instance Learning (MIL) has received heightened research focus as a viable solution for AI-centric diagnostic aid. Recently, to improve its performance and make it work more like a pathologist, several MIL approaches based on the use of multiple-resolution images have been proposed, delivering often higher performance than those that use single-resolution images. Despite impressive recent developments of multiple-resolution MIL, previous approaches only focus on improving performance, thereby lacking research on well-calibrated MIL that clinical experts can rely on for trustworthy diagnostic results. In this study, we propose Uncertainty-Focused Calibrated MIL (UFC-MIL), which more closely mimics the pathologists' examination behaviors while providing calibrated diagnostic predictions, using multiple images with different resolutions. UFC-MIL includes a novel patch-wise loss that learns the latent patterns of instances and expresses their uncertainty for classification. Also, the attention-based architecture with a neighbor patch aggregation module collects features for the classifier. In addition, aggregated predictions are calibrated through patch-level uncertainty without requiring multiple iterative inferences, which is a key practical advantage. Against challenging public datasets, UFC-MIL shows superior performance in model calibration while achieving classification accuracy comparable to that of state-of-the-art methods.
Priority-Aware Pathological Hierarchy Training for Multiple Instance Learning
Jul 28, 2025Abstract:Multiple Instance Learning (MIL) is increasingly being used as a support tool within clinical settings for pathological diagnosis decisions, achieving high performance and removing the annotation burden. However, existing approaches for clinical MIL tasks have not adequately addressed the priority issues that exist in relation to pathological symptoms and diagnostic classes, causing MIL models to ignore priority among classes. To overcome this clinical limitation of MIL, we propose a new method that addresses priority issues using two hierarchies: vertical inter-hierarchy and horizontal intra-hierarchy. The proposed method aligns MIL predictions across each hierarchical level and employs an implicit feature re-usability during training to facilitate clinically more serious classes within the same level. Experiments with real-world patient data show that the proposed method effectively reduces misdiagnosis and prioritizes more important symptoms in multiclass scenarios. Further analysis verifies the efficacy of the proposed components and qualitatively confirms the MIL predictions against challenging cases with multiple symptoms.
STEM image analysis based on deep learning: identification of vacancy defects and polymorphs of ${MoS_2}$
Jun 09, 2022
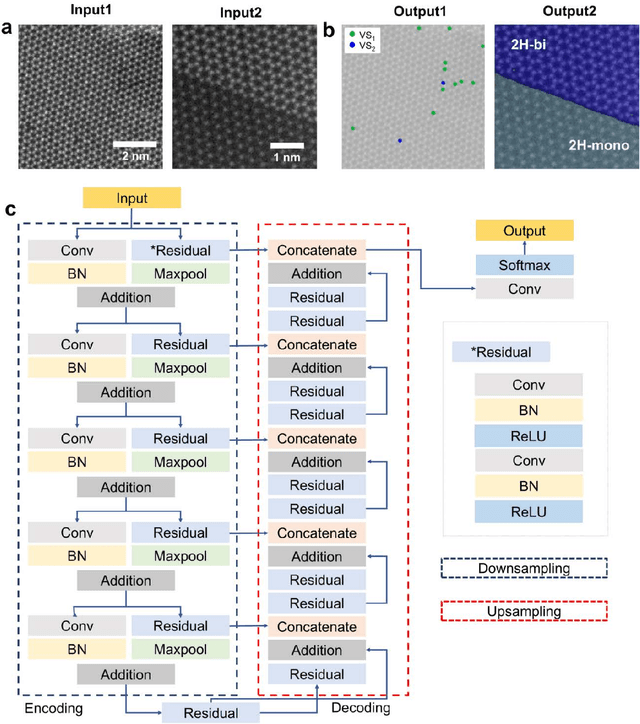
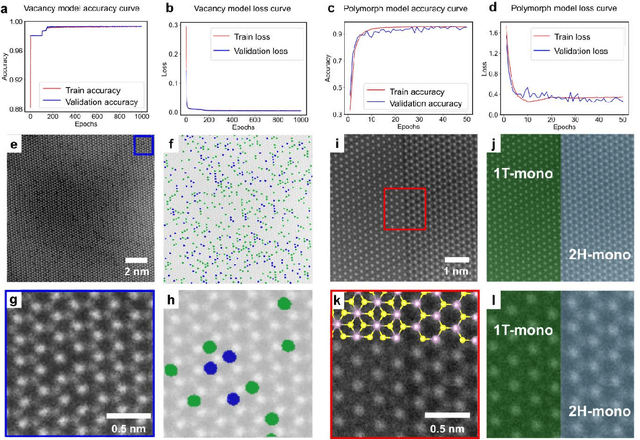
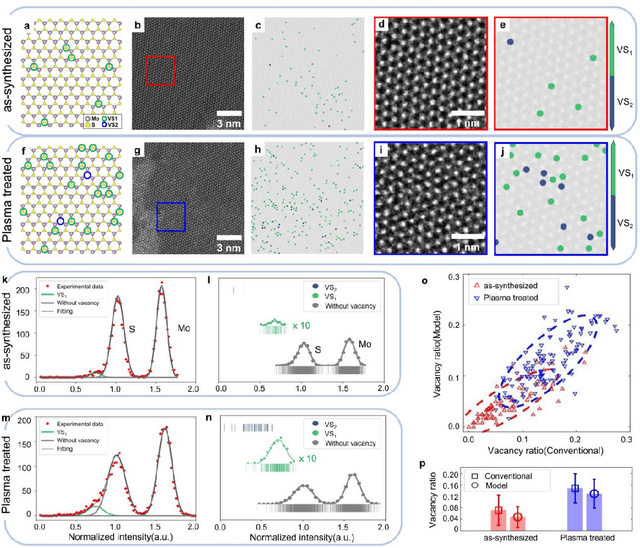
Abstract:Scanning transmission electron microscopy (STEM) is an indispensable tool for atomic-resolution structural analysis for a wide range of materials. The conventional analysis of STEM images is an extensive hands-on process, which limits efficient handling of high-throughput data. Here we apply a fully convolutional network (FCN) for identification of important structural features of two-dimensional crystals. ResUNet, a type of FCN, is utilized in identifying sulfur vacancies and polymorph types of ${MoS_2}$ from atomic resolution STEM images. Efficient models are achieved based on training with simulated images in the presence of different levels of noise, aberrations, and carbon contamination. The accuracy of the FCN models toward extensive experimental STEM images is comparable to that of careful hands-on analysis. Our work provides a guideline on best practices to train a deep learning model for STEM image analysis and demonstrates FCN's application for efficient processing of a large volume of STEM data.
* 24 pages, 5 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge