Shivam Kalra
Pan-Cancer Diagnostic Consensus Through Searching Archival Histopathology Images Using Artificial Intelligence
Nov 20, 2019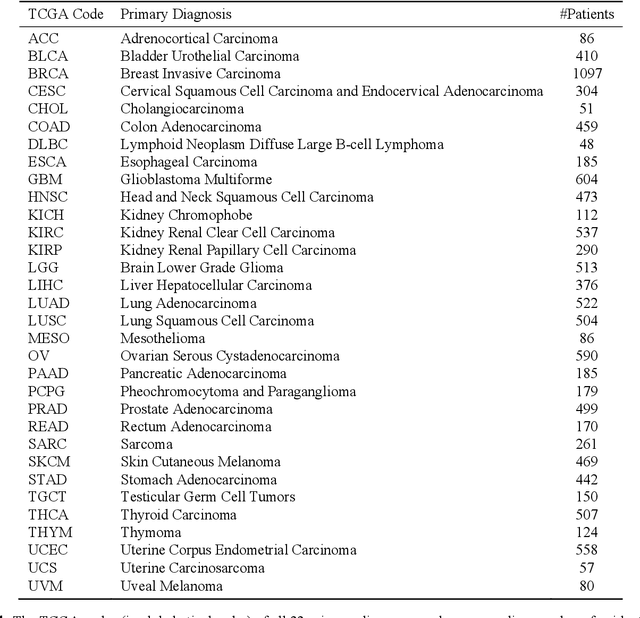
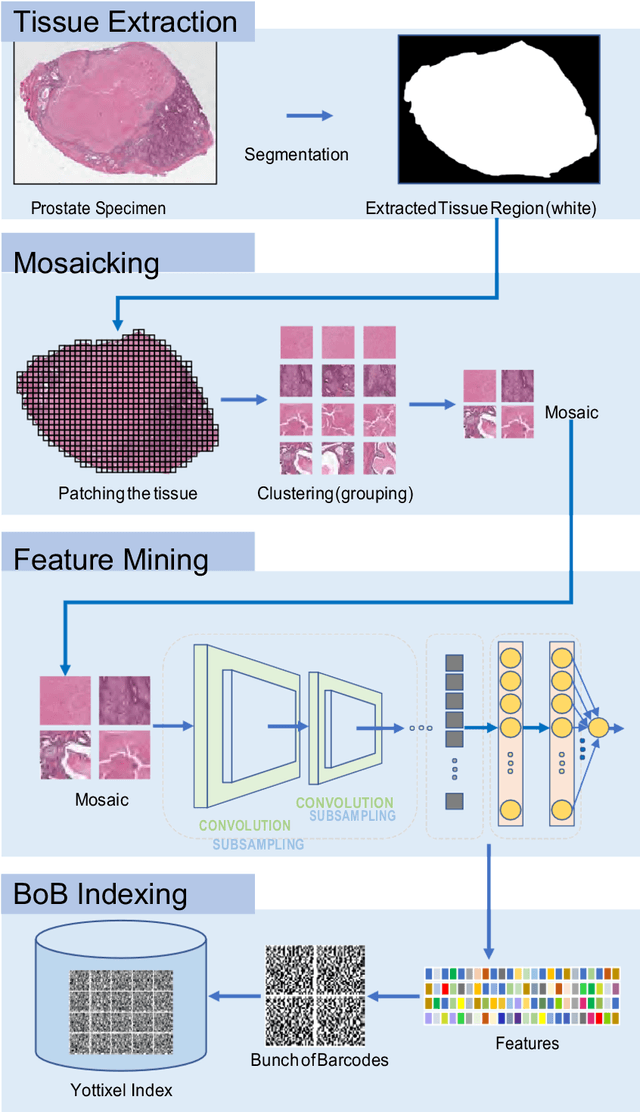
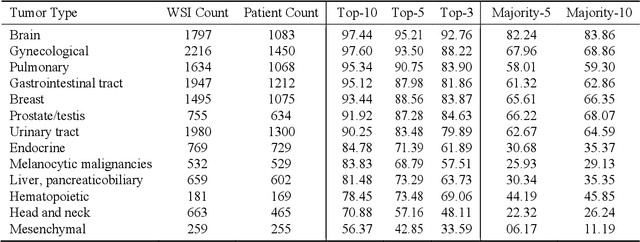
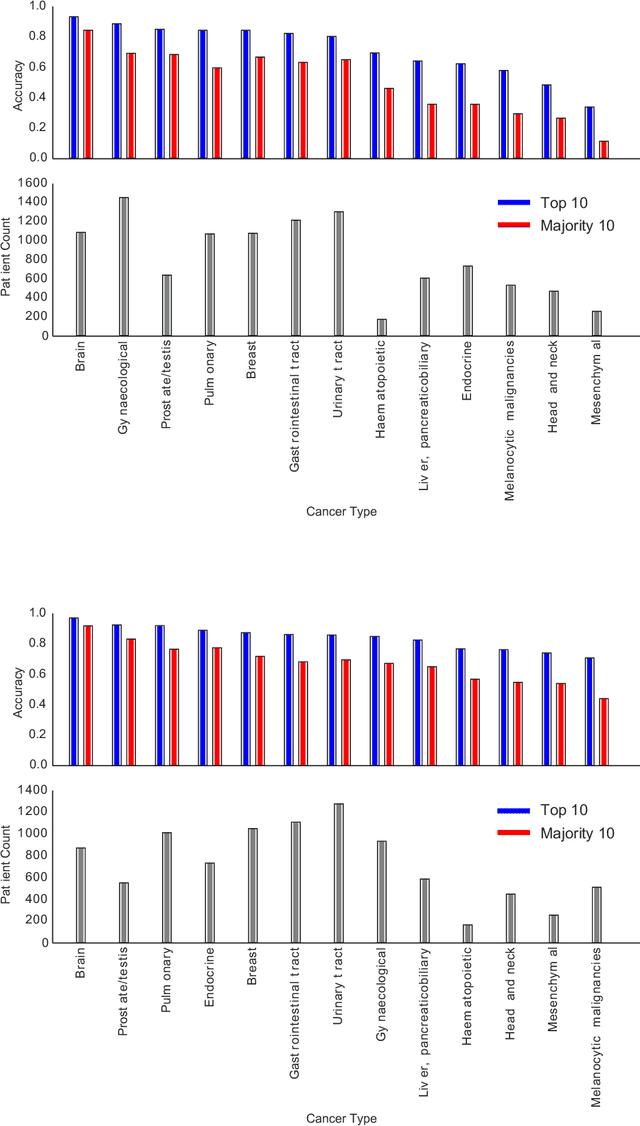
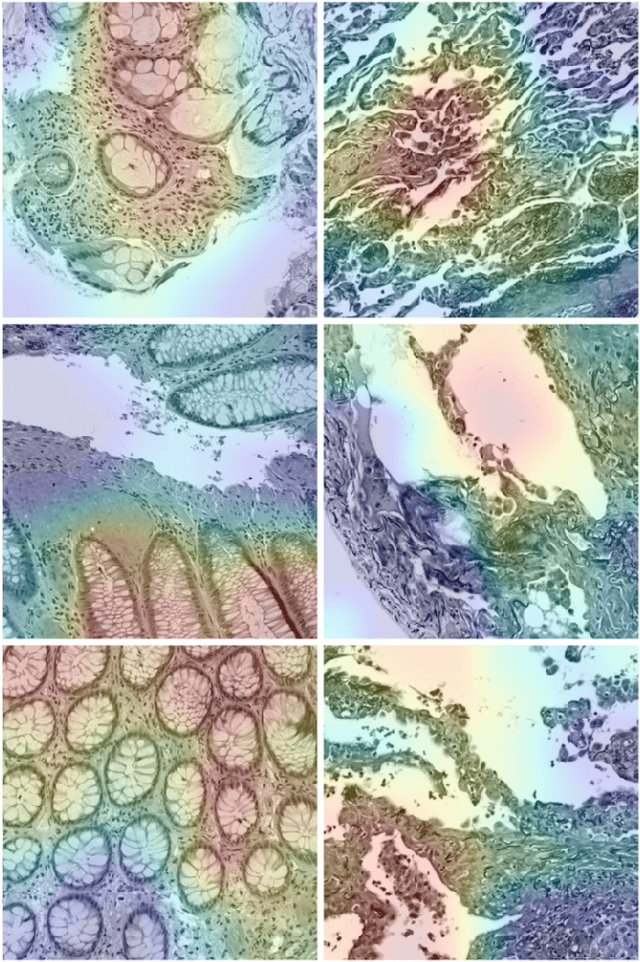
Abstract:The emergence of digital pathology has opened new horizons for histopathology and cytology. Artificial-intelligence algorithms are able to operate on digitized slides to assist pathologists with diagnostic tasks. Whereas machine learning involving classification and segmentation methods have obvious benefits for image analysis in pathology, image search represents a fundamental shift in computational pathology. Matching the pathology of new patients with already diagnosed and curated cases offers pathologist a novel approach to improve diagnostic accuracy through visual inspection of similar cases and computational majority vote for consensus building. In this study, we report the results from searching the largest public repository (The Cancer Genome Atlas [TCGA] program by National Cancer Institute, USA) of whole slide images from almost 11,000 patients depicting different types of malignancies. For the first time, we successfully indexed and searched almost 30,000 high-resolution digitized slides constituting 16 terabytes of data comprised of 20 million 1000x1000 pixels image patches. The TCGA image database covers 25 anatomic sites and contains 32 cancer subtypes. High-performance storage and GPU power were employed for experimentation. The results were assessed with conservative "majority voting" to build consensus for subtype diagnosis through vertical search and demonstrated high accuracy values for both frozen sections slides (e.g., bladder urothelial carcinoma 93%, kidney renal clear cell carcinoma 97%, and ovarian serous cystadenocarcinoma 99%) and permanent histopathology slides (e.g., prostate adenocarcinoma 98%, skin cutaneous melanoma 99%, and thymoma 100%). The key finding of this validation study was that computational consensus appears to be possible for rendering diagnoses if a sufficiently large number of searchable cases are available for each cancer subtype.
Learning Permutation Invariant Representations using Memory Networks
Nov 18, 2019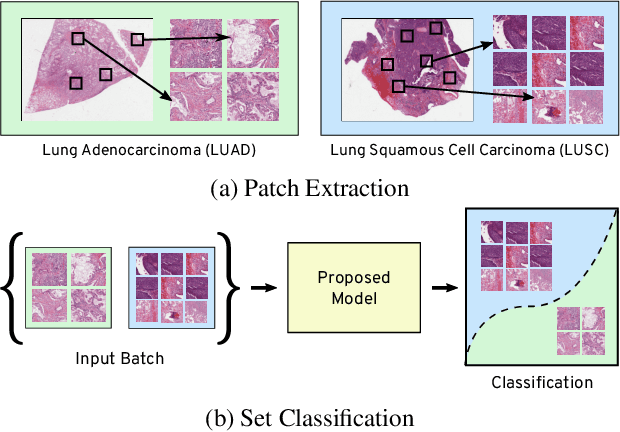
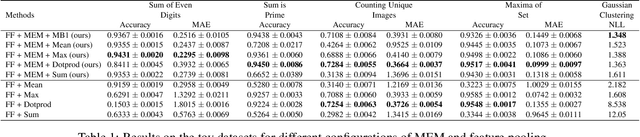
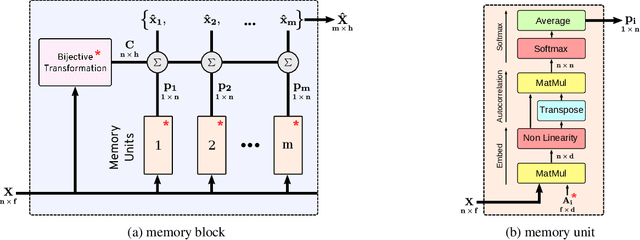

Abstract:Many real world tasks such as 3D object detection and high-resolution image classification involve learning from a set of instances. In these cases, only a group of instances, a set, collectively contains meaningful information and therefore only the sets have labels, and not individual data instances. In this work, we present a permutation invariant neural network called a \textbf{Memory-based Exchangeable Model (MEM)} for learning set functions. The model consists of memory units that embed an input sequence to high-level features (memories) enabling the model to learn inter-dependencies among instances of the set in the form of attention vectors. To demonstrate its learning ability, we evaluated our model on test datasets created using MNIST, point cloud classification, and population estimation. We also tested the model for classifying histopathology whole slide images to discriminate between two subtypes of Lung cancer---Lung Adenocarcinoma, and Lung Squamous Cell Carcinoma. We systematically extracted patches from lung cancer images from The Cancer Genome Atlas~(TCGA) dataset, the largest public repository of histopathology images. The proposed method achieved a competitive classification accuracy of 84.84\%. The results on other datasets are promising and demonstrate the efficacy of our model.
Subtractive Perceptrons for Learning Images: A Preliminary Report
Sep 15, 2019
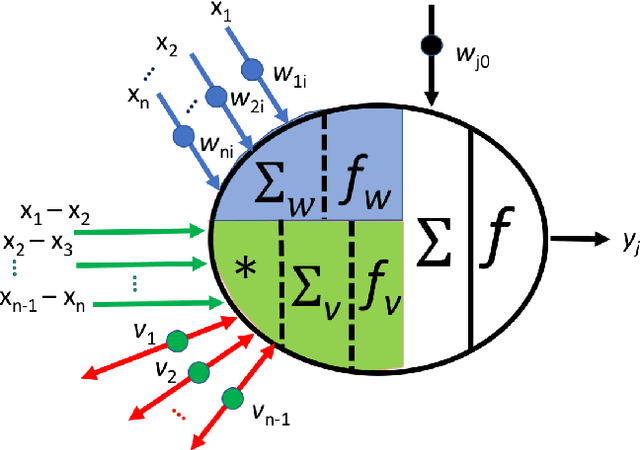
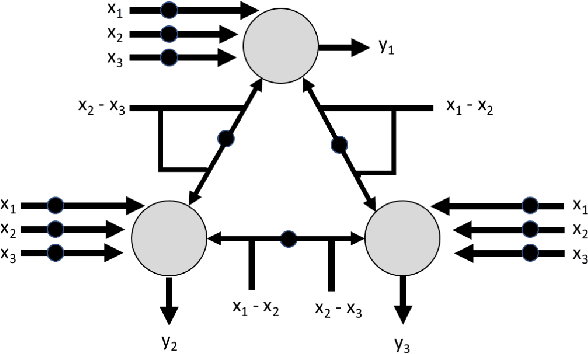
Abstract:In recent years, artificial neural networks have achieved tremendous success for many vision-based tasks. However, this success remains within the paradigm of \emph{weak AI} where networks, among others, are specialized for just one given task. The path toward \emph{strong AI}, or Artificial General Intelligence, remains rather obscure. One factor, however, is clear, namely that the feed-forward structure of current networks is not a realistic abstraction of the human brain. In this preliminary work, some ideas are proposed to define a \textit{subtractive Perceptron} (s-Perceptron), a graph-based neural network that delivers a more compact topology to learn one specific task. In this preliminary study, we test the s-Perceptron with the MNIST dataset, a commonly used image archive for digit recognition. The proposed network achieves excellent results compared to the benchmark networks that rely on more complex topologies.
Projectron -- A Shallow and Interpretable Network for Classifying Medical Images
Mar 15, 2019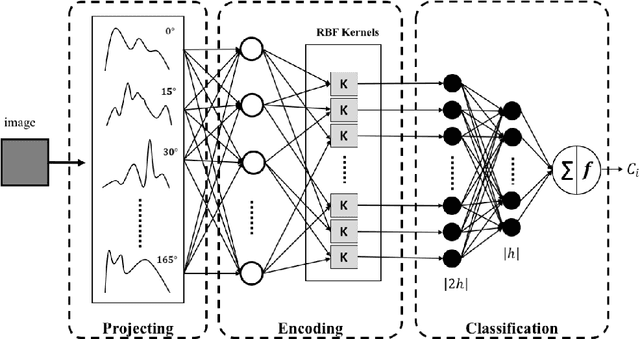
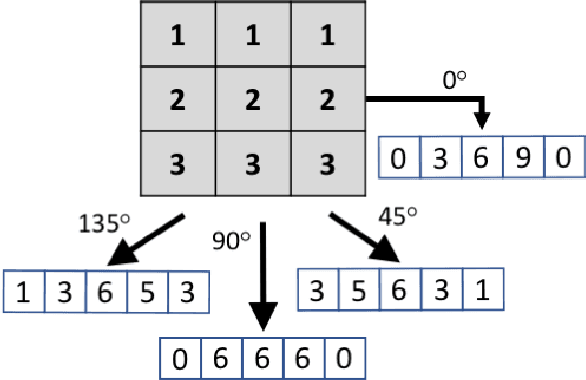
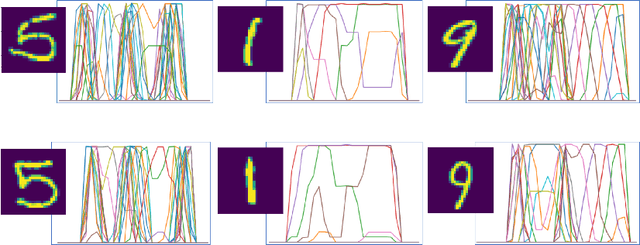
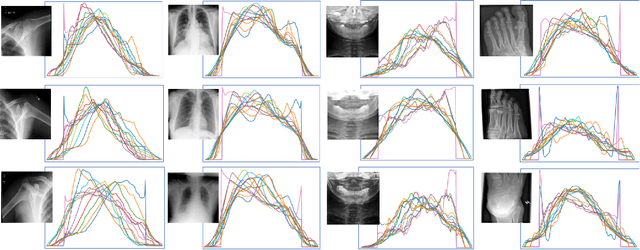
Abstract:This paper introduces the `Projectron' as a new neural network architecture that uses Radon projections to both classify and represent medical images. The motivation is to build shallow networks which are more interpretable in the medical imaging domain. Radon transform is an established technique that can reconstruct images from parallel projections. The Projectron first applies global Radon transform to each image using equidistant angles and then feeds these transformations for encoding to a single layer of neurons followed by a layer of suitable kernels to facilitate a linear separation of projections. Finally, the Projectron provides the output of the encoding as an input to two more layers for final classification. We validate the Projectron on five publicly available datasets, a general dataset (namely MNIST) and four medical datasets (namely Emphysema, IDC, IRMA, and Pneumonia). The results are encouraging as we compared the Projectron's performance against MLPs with raw images and Radon projections as inputs, respectively. Experiments clearly demonstrate the potential of the proposed Projectron for representing/classifying medical images.
Automatic Classification of Pathology Reports using TF-IDF Features
Mar 05, 2019
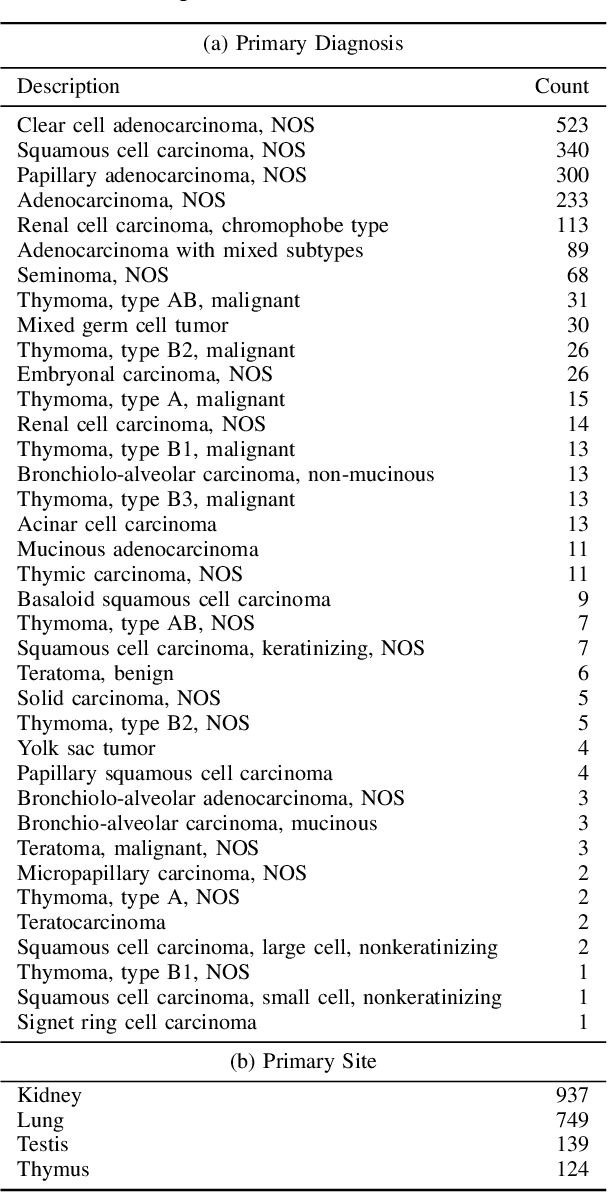
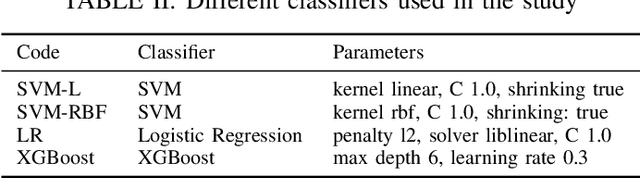
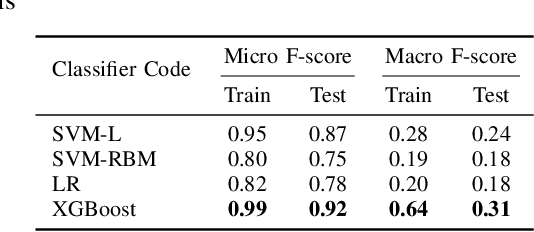
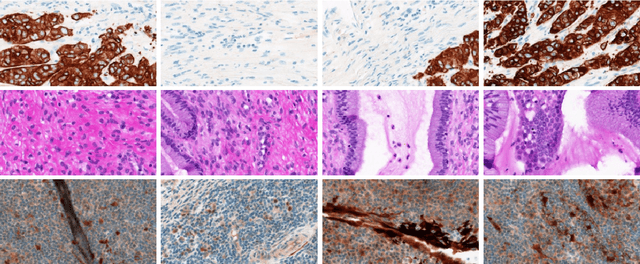
Abstract:A Pathology report is arguably one of the most important documents in medicine containing interpretive information about the visual findings from the patient's biopsy sample. Each pathology report has a retention period of up to 20 years after the treatment of a patient. Cancer registries process and encode high volumes of free-text pathology reports for surveillance of cancer and tumor diseases all across the world. In spite of their extremely valuable information they hold, pathology reports are not used in any systematic way to facilitate computational pathology. Therefore, in this study, we investigate automated machine-learning techniques to identify/predict the primary diagnosis (based on ICD-O code) from pathology reports. We performed experiments by extracting the TF-IDF features from the reports and classifying them using three different methods---SVM, XGBoost, and Logistic Regression. We constructed a new dataset with 1,949 pathology reports arranged into 37 ICD-O categories, collected from four different primary sites, namely lung, kidney, thymus, and testis. The reports were manually transcribed into text format after collecting them as PDF files from NCI Genomic Data Commons public dataset. We subsequently pre-processed the reports by removing irrelevant textual artifacts produced by OCR software. The highest classification accuracy we achieved was 92\% using XGBoost classifier on TF-IDF feature vectors, the linear SVM scored 87\% accuracy. Furthermore, the study shows that TF-IDF vectors are suitable for highlighting the important keywords within a report which can be helpful for the cancer research and diagnostic workflow. The results are encouraging in demonstrating the potential of machine learning methods for classification and encoding of pathology reports.
Convolutional Neural Networks for Histopathology Image Classification: Training vs. Using Pre-Trained Networks
Oct 11, 2017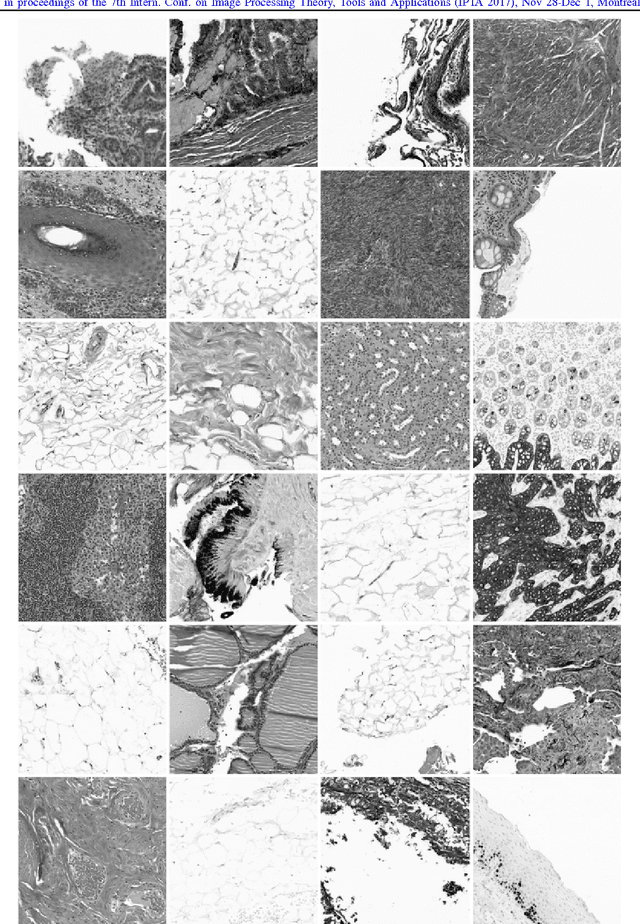

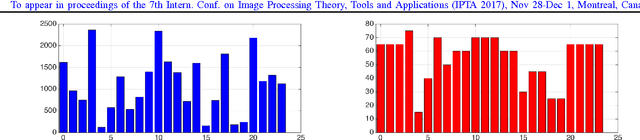
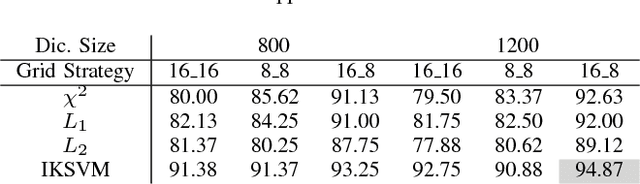
Abstract:We explore the problem of classification within a medical image data-set based on a feature vector extracted from the deepest layer of pre-trained Convolution Neural Networks. We have used feature vectors from several pre-trained structures, including networks with/without transfer learning to evaluate the performance of pre-trained deep features versus CNNs which have been trained by that specific dataset as well as the impact of transfer learning with a small number of samples. All experiments are done on Kimia Path24 dataset which consists of 27,055 histopathology training patches in 24 tissue texture classes along with 1,325 test patches for evaluation. The result shows that pre-trained networks are quite competitive against training from scratch. As well, fine-tuning does not seem to add any tangible improvement for VGG16 to justify additional training while we observed considerable improvement in retrieval and classification accuracy when we fine-tuned the Inception structure.
A Comparative Study of CNN, BoVW and LBP for Classification of Histopathological Images
Sep 27, 2017
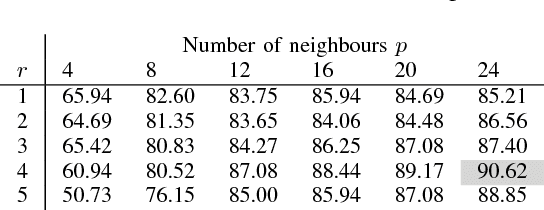
Abstract:Despite the progress made in the field of medical imaging, it remains a large area of open research, especially due to the variety of imaging modalities and disease-specific characteristics. This paper is a comparative study describing the potential of using local binary patterns (LBP), deep features and the bag-of-visual words (BoVW) scheme for the classification of histopathological images. We introduce a new dataset, \emph{KIMIA Path960}, that contains 960 histopathology images belonging to 20 different classes (different tissue types). We make this dataset publicly available. The small size of the dataset and its inter- and intra-class variability makes it ideal for initial investigations when comparing image descriptors for search and classification in complex medical imaging cases like histopathology. We investigate deep features, LBP histograms and BoVW to classify the images via leave-one-out validation. The accuracy of image classification obtained using LBP was 90.62\% while the highest accuracy using deep features reached 94.72\%. The dictionary approach (BoVW) achieved 96.50\%. Deep solutions may be able to deliver higher accuracies but they need extensive training with a large number of (balanced) image datasets.
Skin Lesion Segmentation: U-Nets versus Clustering
Sep 27, 2017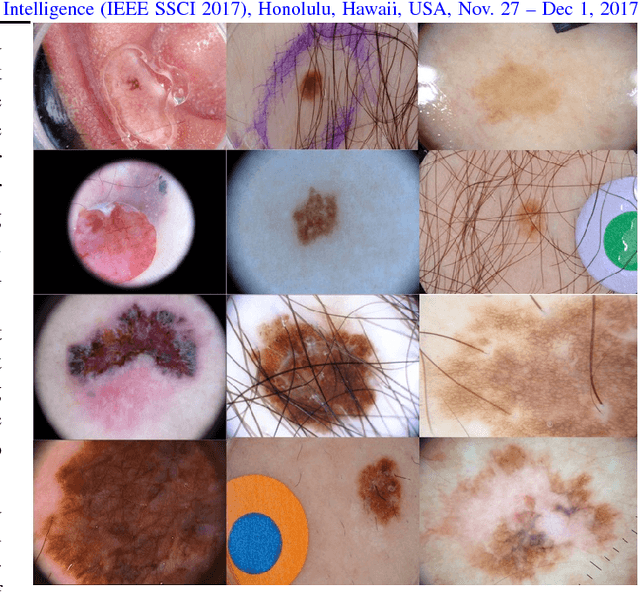


Abstract:Many automatic skin lesion diagnosis systems use segmentation as a preprocessing step to diagnose skin conditions because skin lesion shape, border irregularity, and size can influence the likelihood of malignancy. This paper presents, examines and compares two different approaches to skin lesion segmentation. The first approach uses U-Nets and introduces a histogram equalization based preprocessing step. The second approach is a C-Means clustering based approach that is much simpler to implement and faster to execute. The Jaccard Index between the algorithm output and hand segmented images by dermatologists is used to evaluate the proposed algorithms. While many recently proposed deep neural networks to segment skin lesions require a significant amount of computational power for training (i.e., computer with GPUs), the main objective of this paper is to present methods that can be used with only a CPU. This severely limits, for example, the number of training instances that can be presented to the U-Net. Comparing the two proposed algorithms, U-Nets achieved a significantly higher Jaccard Index compared to the clustering approach. Moreover, using the histogram equalization for preprocessing step significantly improved the U-Net segmentation results.
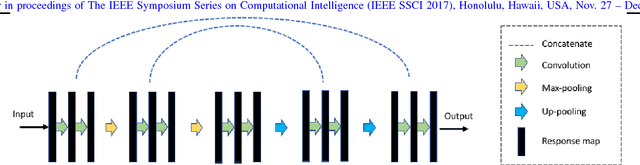
Learning Autoencoded Radon Projections
Sep 27, 2017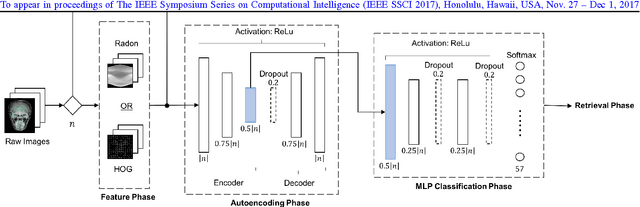
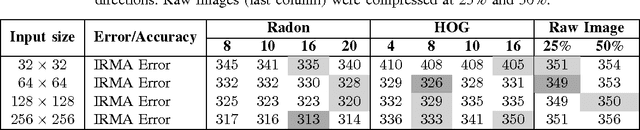
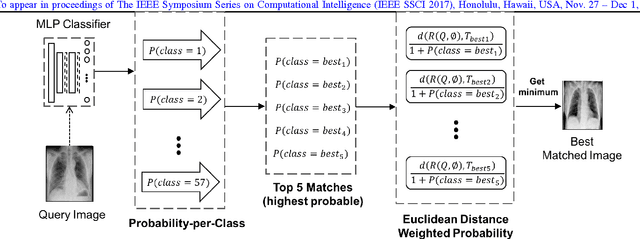
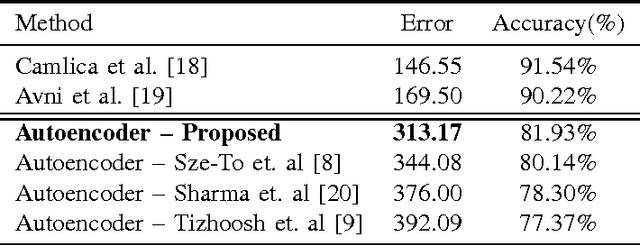
Abstract:Autoencoders have been recently used for encoding medical images. In this study, we design and validate a new framework for retrieving medical images by classifying Radon projections, compressed in the deepest layer of an autoencoder. As the autoencoder reduces the dimensionality, a multilayer perceptron (MLP) can be employed to classify the images. The integration of MLP promotes a rather shallow learning architecture which makes the training faster. We conducted a comparative study to examine the capabilities of autoencoders for different inputs such as raw images, Histogram of Oriented Gradients (HOG) and normalized Radon projections. Our framework is benchmarked on IRMA dataset containing $14,410$ x-ray images distributed across $57$ different classes. Experiments show an IRMA error of $313$ (equivalent to $\approx 82\%$ accuracy) outperforming state-of-the-art works on retrieval from IRMA dataset using autoencoders.
Classification and Retrieval of Digital Pathology Scans: A New Dataset
May 22, 2017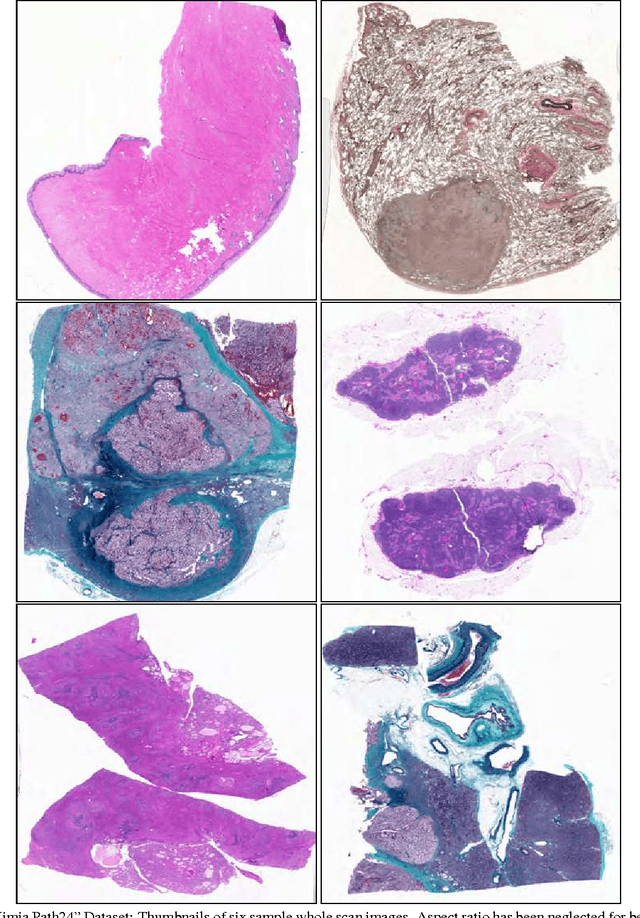


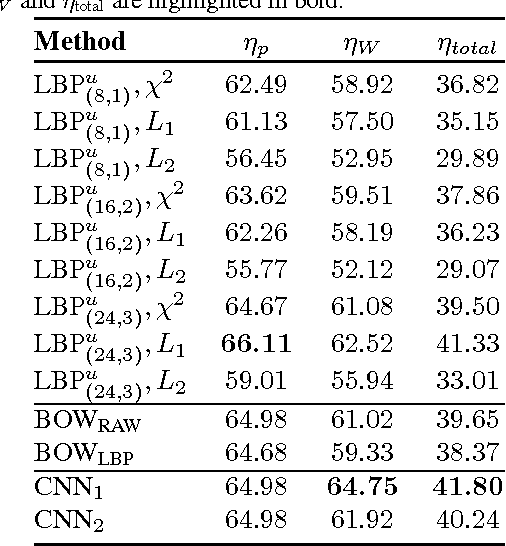
Abstract:In this paper, we introduce a new dataset, \textbf{Kimia Path24}, for image classification and retrieval in digital pathology. We use the whole scan images of 24 different tissue textures to generate 1,325 test patches of size 1000$\times$1000 (0.5mm$\times$0.5mm). Training data can be generated according to preferences of algorithm designer and can range from approximately 27,000 to over 50,000 patches if the preset parameters are adopted. We propose a compound patch-and-scan accuracy measurement that makes achieving high accuracies quite challenging. In addition, we set the benchmarking line by applying LBP, dictionary approach and convolutional neural nets (CNNs) and report their results. The highest accuracy was 41.80\% for CNN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge