Sergei Grudinin
DAO
BAnG: Bidirectional Anchored Generation for Conditional RNA Design
Feb 28, 2025Abstract:Designing RNA molecules that interact with specific proteins is a critical challenge in experimental and computational biology. Existing computational approaches require a substantial amount of experimentally determined RNA sequences for each specific protein or a detailed knowledge of RNA structure, restricting their utility in practice. To address this limitation, we develop RNA-BAnG, a deep learning-based model designed to generate RNA sequences for protein interactions without these requirements. Central to our approach is a novel generative method, Bidirectional Anchored Generation (BAnG), which leverages the observation that protein-binding RNA sequences often contain functional binding motifs embedded within broader sequence contexts. We first validate our method on generic synthetic tasks involving similar localized motifs to those appearing in RNAs, demonstrating its benefits over existing generative approaches. We then evaluate our model on biological sequences, showing its effectiveness for conditional RNA sequence design given a binding protein.
From Epilepsy Seizures Classification to Detection: A Deep Learning-based Approach for Raw EEG Signals
Oct 04, 2024Abstract:Epilepsy represents the most prevalent neurological disease in the world. One-third of people suffering from mesial temporal lobe epilepsy (MTLE) exhibit drug resistance, urging the need to develop new treatments. A key part in anti-seizure medication (ASM) development is the capability of detecting and quantifying epileptic seizures occurring in electroencephalogram (EEG) signals, which is crucial for treatment efficacy evaluation. In this study, we introduced a seizure detection pipeline based on deep learning models applied to raw EEG signals. This pipeline integrates: a new pre-processing technique which segments continuous raw EEG signals without prior distinction between seizure and seizure-free activities; a post-processing algorithm developed to reassemble EEG segments and allow the identification of seizures start/end; and finally, a new evaluation procedure based on a strict seizure events comparison between predicted and real labels. Models training have been performed using a data splitting strategy which addresses the potential for data leakage. We demonstrated the fundamental differences between a seizure classification and a seizure detection task and showed the differences in performance between the two tasks. Finally, we demonstrated the generalization capabilities across species of our best architecture, combining a Convolutional Neural Network and a Transformer encoder. The model was trained on animal EEGs and tested on human EEGs with a F1-score of 93% on a balanced Bonn dataset.
On the Fourier analysis in the SO space : EquiLoPO Network
Apr 24, 2024Abstract:Analyzing volumetric data with rotational invariance or equivariance is an active topic in current research. Existing deep-learning approaches utilize either group convolutional networks limited to discrete rotations or steerable convolutional networks with constrained filter structures. This work proposes a novel equivariant neural network architecture that achieves analytical Equivariance to Local Pattern Orientation on the continuous SO(3) group while allowing unconstrained trainable filters - EquiLoPO Network. Our key innovations are a group convolutional operation leveraging irreducible representations as the Fourier basis and a local activation function in the SO(3) space that provides a well-defined mapping from input to output functions, preserving equivariance. By integrating these operations into a ResNet-style architecture, we propose a model that overcomes the limitations of prior methods. A comprehensive evaluation on diverse 3D medical imaging datasets from MedMNIST3D demonstrates the effectiveness of our approach, which consistently outperforms state of the art. This work suggests the benefits of true rotational equivariance on SO(3) and flexible unconstrained filters enabled by the local activation function, providing a flexible framework for equivariant deep learning on volumetric data with potential applications across domains. Our code is publicly available at \url{https://gricad-gitlab.univ-grenoble-alpes.fr/GruLab/ILPO/-/tree/main/EquiLoPO}.
ILPO-NET: Network for the invariant recognition of arbitrary volumetric patterns in 3D
Apr 04, 2024Abstract:Effective recognition of spatial patterns and learning their hierarchy is crucial in modern spatial data analysis. Volumetric data applications seek techniques ensuring invariance not only to shifts but also to pattern rotations. While traditional methods can readily achieve translational invariance, rotational invariance possesses multiple challenges and remains an active area of research. Here, we present ILPO-Net (Invariant to Local Patterns Orientation Network), a novel approach that handles arbitrarily shaped patterns with the convolutional operation inherently invariant to local spatial pattern orientations using the Wigner matrix expansions. Our architecture seamlessly integrates the new convolution operator and, when benchmarked on diverse volumetric datasets such as MedMNIST and CATH, demonstrates superior performance over the baselines with significantly reduced parameter counts - up to 1000 times fewer in the case of MedMNIST. Beyond these demonstrations, ILPO-Net's rotational invariance paves the way for other applications across multiple disciplines. Our code is publicly available at https://gricad-gitlab.univ-grenoble-alpes.fr/GruLab/ILPONet.
6DCNN with roto-translational convolution filters for volumetric data processing
Jul 30, 2021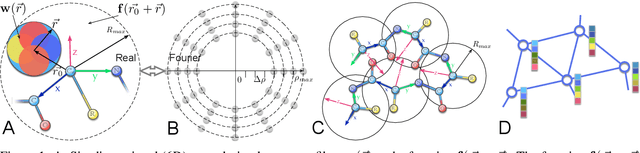
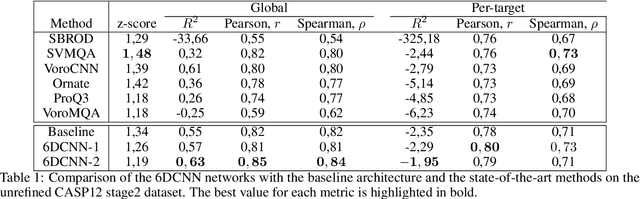
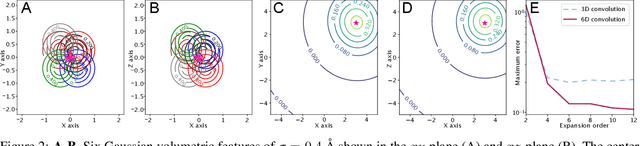
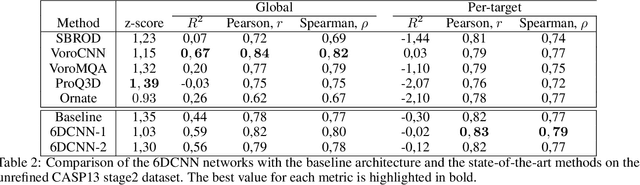
Abstract:In this work, we introduce 6D Convolutional Neural Network (6DCNN) designed to tackle the problem of detecting relative positions and orientations of local patterns when processing three-dimensional volumetric data. 6DCNN also includes SE(3)-equivariant message-passing and nonlinear activation operations constructed in the Fourier space. Working in the Fourier space allows significantly reducing the computational complexity of our operations. We demonstrate the properties of the 6D convolution and its efficiency in the recognition of spatial patterns. We also assess the 6DCNN model on several datasets from the recent CASP protein structure prediction challenges. Here, 6DCNN improves over the baseline architecture and also outperforms the state of the art.
Protein sequence-to-structure learning: Is this the end(-to-end revolution)?
May 16, 2021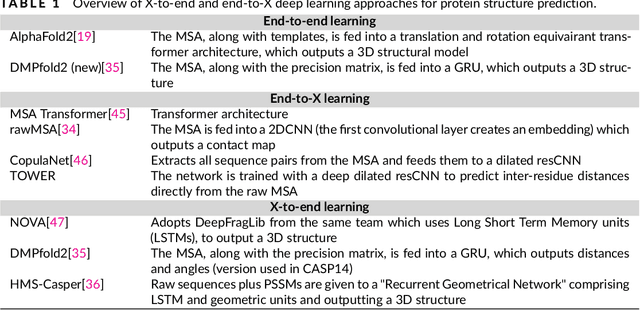
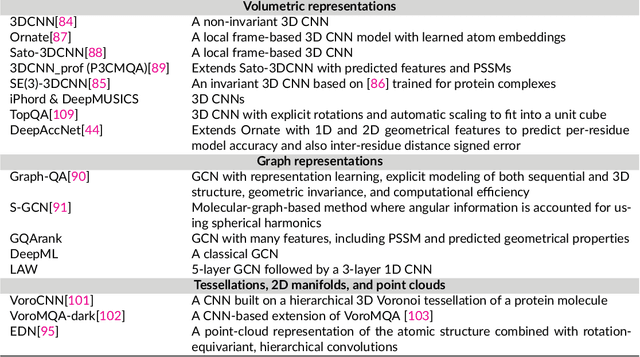
Abstract:The potential of deep learning has been recognized in the protein structure prediction community for some time, and became indisputable after CASP13. In CASP14, deep learning has boosted the field to unanticipated levels reaching near-experimental accuracy. This success comes from advances transferred from other machine learning areas, as well as methods specifically designed to deal with protein sequences and structures, and their abstractions. Novel emerging approaches include (i) geometric learning, i.e. learning on representations such as graphs, 3D Voronoi tessellations, and point clouds; (ii) pre-trained protein language models leveraging attention; (iii) equivariant architectures preserving the symmetry of 3D space; (iv) use of large meta-genome databases; (v) combinations of protein representations; (vi) and finally truly end-to-end architectures, i.e. differentiable models starting from a sequence and returning a 3D structure. Here, we provide an overview and our opinion of the novel deep learning approaches developed in the last two years and widely used in CASP14.
Spherical convolutions on molecular graphs for protein model quality assessment
Nov 16, 2020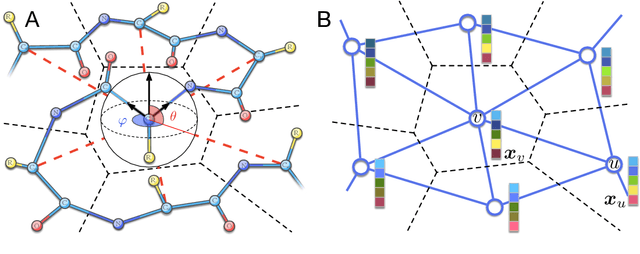
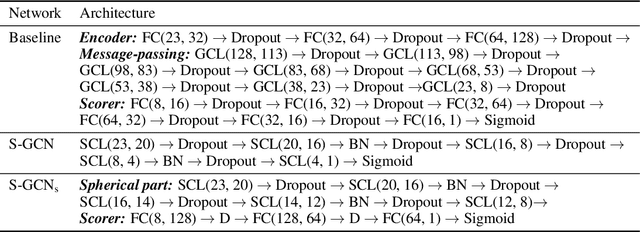
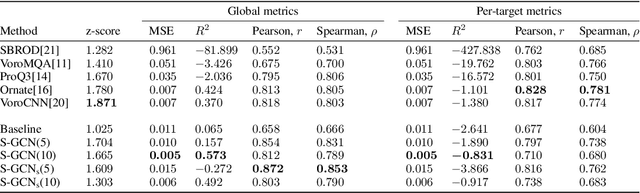
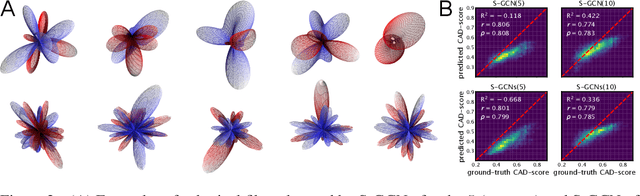
Abstract:Processing information on 3D objects requires methods stable to rigid-body transformations, in particular rotations, of the input data. In image processing tasks, convolutional neural networks achieve this property using rotation-equivariant operations. However, contrary to images, graphs generally have irregular topology. This makes it challenging to define a rotation-equivariant convolution operation on these structures. In this work, we propose Spherical Graph Convolutional Network (S-GCN) that processes 3D models of proteins represented as molecular graphs. In a protein molecule, individual amino acids have common topological elements. This allows us to unambiguously associate each amino acid with a local coordinate system and construct rotation-equivariant spherical filters that operate on angular information between graph nodes. Within the framework of the protein model quality assessment problem, we demonstrate that the proposed spherical convolution method significantly improves the quality of model assessment compared to the standard message-passing approach. It is also comparable to state-of-the-art methods, as we demonstrate on Critical Assessment of Structure Prediction (CASP) benchmarks. The proposed technique operates only on geometric features of protein 3D models. This makes it universal and applicable to any other geometric-learning task where the graph structure allows constructing local coordinate systems.
DeepSymmetry : Using 3D convolutional networks for identification of tandem repeats and internal symmetries in protein structures
Oct 29, 2018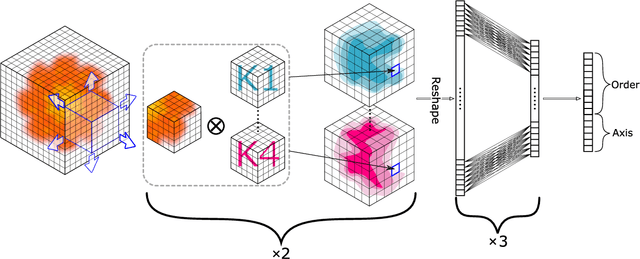

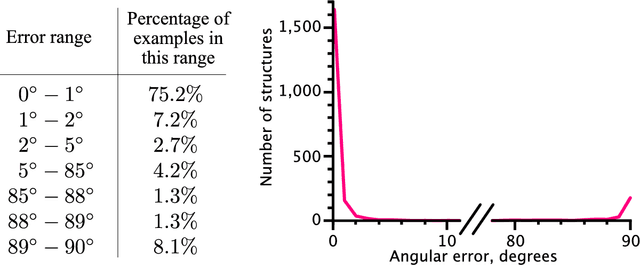
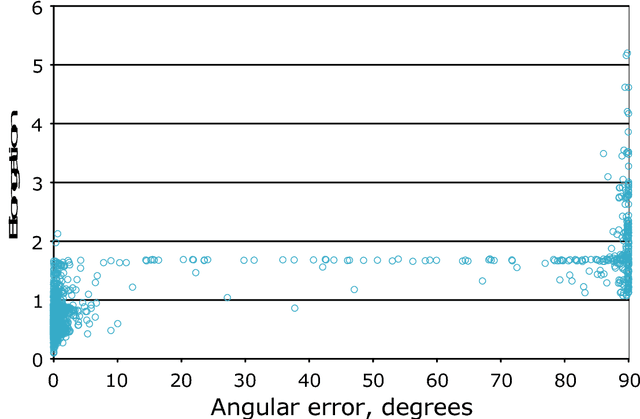
Abstract:Motivation: Thanks to the recent advances in structural biology, nowadays three-dimensional structures of various proteins are solved on a routine basis. A large portion of these contain structural repetitions or internal symmetries. To understand the evolution mechanisms of these proteins and how structural repetitions affect the protein function, we need to be able to detect such proteins very robustly. As deep learning is particularly suited to deal with spatially organized data, we applied it to the detection of proteins with structural repetitions. Results: We present DeepSymmetry, a versatile method based on three-dimensional (3D) convolutional networks that detects structural repetitions in proteins and their density maps. Our method is designed to identify tandem repeat proteins, proteins with internal symmetries, symmetries in the raw density maps, their symmetry order, and also the corresponding symmetry axes. Detection of symmetry axes is based on learning six-dimensional Veronese mappings of 3D vectors, and the median angular error of axis determination is less than one degree. We demonstrate the capabilities of our method on benchmarks with tandem repeated proteins and also with symmetrical assemblies. For example, we have discovered over 10,000 putative tandem repeat proteins that are not currently present in the RepeatsDB database. Availability: The method is available at https://team.inria.fr/nano-d/software/deepsymmetry. It consists of a C++ executable that transforms molecular structures into volumetric density maps, and a Python code based on the TensorFlow framework for applying the DeepSymmetry model to these maps.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge