Sebastian Berisha
Rapid hyperspectral photothermal mid-infrared spectroscopic imaging from sparse data for gynecologic cancer tissue subtyping
Feb 28, 2024



Abstract:Ovarian cancer detection has traditionally relied on a multi-step process that includes biopsy, tissue staining, and morphological analysis by experienced pathologists. While widely practiced, this conventional approach suffers from several drawbacks: it is qualitative, time-intensive, and heavily dependent on the quality of staining. Mid-infrared (MIR) hyperspectral photothermal imaging is a label-free, biochemically quantitative technology that, when combined with machine learning algorithms, can eliminate the need for staining and provide quantitative results comparable to traditional histology. However, this technology is slow. This work presents a novel approach to MIR photothermal imaging that enhances its speed by an order of magnitude. Our method significantly accelerates data collection by capturing a combination of high-resolution and interleaved, lower-resolution infrared band images and applying computational techniques for data interpolation. We effectively minimize data collection requirements by leveraging sparse data acquisition and employing curvelet-based reconstruction algorithms. This method enables the reconstruction of high-quality, high-resolution images from undersampled datasets and achieving a 10X improvement in data acquisition time. We assessed the performance of our sparse imaging methodology using a variety of quantitative metrics, including mean squared error (MSE), structural similarity index (SSIM), and tissue subtype classification accuracies, employing both random forest and convolutional neural network (CNN) models, accompanied by ROC curves. Our statistically robust analysis, based on data from 100 ovarian cancer patient samples and over 65 million data points, demonstrates the method's capability to produce superior image quality and accurately distinguish between different gynecological tissue types with segmentation accuracy exceeding 95%.
Leveraging high-resolution spatial features in mid-infrared spectroscopic imaging to classify tissue subtypes in ovarian cancer
May 19, 2022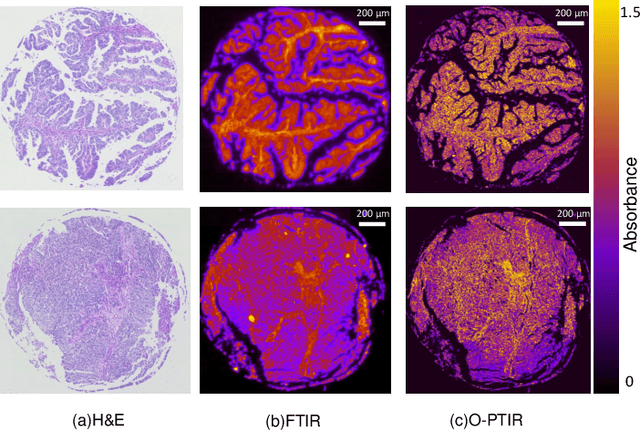

Abstract:Mid-infrared spectroscopic imaging (MIRSI) is an emerging class of label-free techniques being leveraged for digital histopathology. Optical photothermal infrared (O-PTIR) is based on vibrational absorbance imaging using a pump-probe architecture capable of a 10x enhancement in spatial resolution relative to FTIR imaging. This allows truly sub-cellular spectroscopic investigation of tissue at biochemically important fingerprint wavelengths. Modern histopathologic identification of ovarian cancer involves tissue staining followed by morphological pattern recognition. This process is time-consuming, subjective, and requires extensive expertise. In this paper, we present the first label-free automated histological classification of ovarian tissue sub-types using MIRSI. We demonstrate that enhanced resolution of sub-cellular features, combined with spectroscopic information, enables reliable classification (0.98 AUC) of ovarian cell sub-types. Moreover, we present statistically robust validation from 74 patient samples with over 60 million data points. This demonstrates that sub-cellular resolution from five wavenumbers is sufficient to outperform state-of-the-art diffraction-limited techniques from up to 374 different wavenumbers. O-PTIR also performs measurements in back-reflection geometry, opening the door to future in vivo studies on glass slides.
Three-Dimensional GPU-Accelerated Active Contours for Automated Localization of Cells in Large Images
Apr 17, 2018
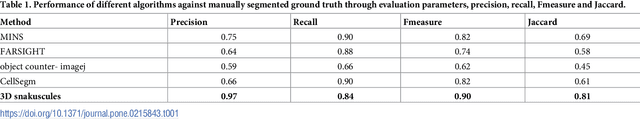
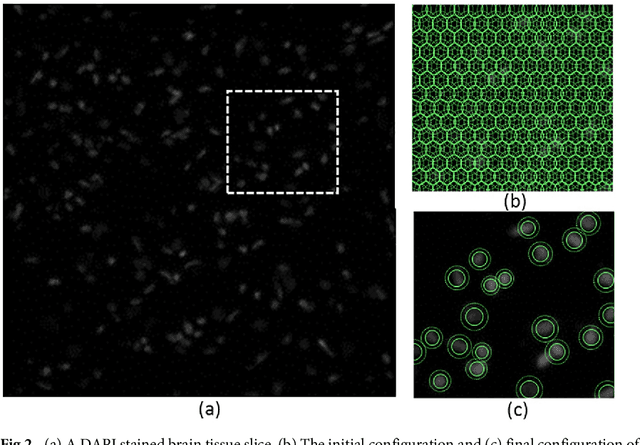

Abstract:Cell segmentation in microscopy is a challenging problem, since cells are often asymmetric and densely packed. This becomes particularly challenging for extremely large images, since manual intervention and processing time can make segmentation intractable. In this paper, we present an efficient and highly parallel formulation for symmetric three-dimensional (3D) contour evolution that extends previous work on fast two-dimensional active contours. We provide a formulation for optimization on 3D images, as well as a strategy for accelerating computation on consumer graphics hardware. The proposed software takes advantage of Monte-Carlo sampling schemes in order to speed up convergence and reduce thread divergence. Experimental results show that this method provides superior performance for large 2D and 3D cell segmentation tasks when compared to existing methods on large 3D brain images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge