Satish Viswanath
Developing Predictive and Robust Radiomics Models for Chemotherapy Response in High-Grade Serous Ovarian Carcinoma
Jan 13, 2026Abstract:Objectives: High-grade serous ovarian carcinoma (HGSOC) is typically diagnosed at an advanced stage with extensive peritoneal metastases, making treatment challenging. Neoadjuvant chemotherapy (NACT) is often used to reduce tumor burden before surgery, but about 40% of patients show limited response. Radiomics, combined with machine learning (ML), offers a promising non-invasive method for predicting NACT response by analyzing computed tomography (CT) imaging data. This study aimed to improve response prediction in HGSOC patients undergoing NACT by integration different feature selection methods. Materials and methods: A framework for selecting robust radiomics features was introduced by employing an automated randomisation algorithm to mimic inter-observer variability, ensuring a balance between feature robustness and prediction accuracy. Four response metrics were used: chemotherapy response score (CRS), RECIST, volume reduction (VolR), and diameter reduction (DiaR). Lesions in different anatomical sites were studied. Pre- and post-NACT CT scans were used for feature extraction and model training on one cohort, and an independent cohort was used for external testing. Results: The best prediction performance was achieved using all lesions combined for VolR prediction, with an AUC of 0.83. Omental lesions provided the best results for CRS prediction (AUC 0.77), while pelvic lesions performed best for DiaR (AUC 0.76). Conclusion: The integration of robustness into the feature selection processes ensures the development of reliable models and thus facilitates the implementation of the radiomics models in clinical applications for HGSOC patients. Future work should explore further applications of radiomics in ovarian cancer, particularly in real-time clinical settings.
Correlation between image quality metrics of magnetic resonance images and the neural network segmentation accuracy
Nov 01, 2021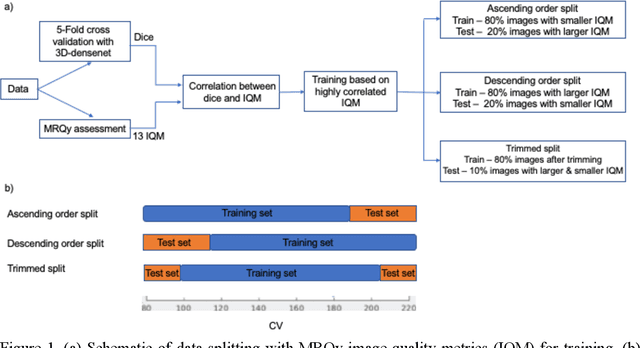
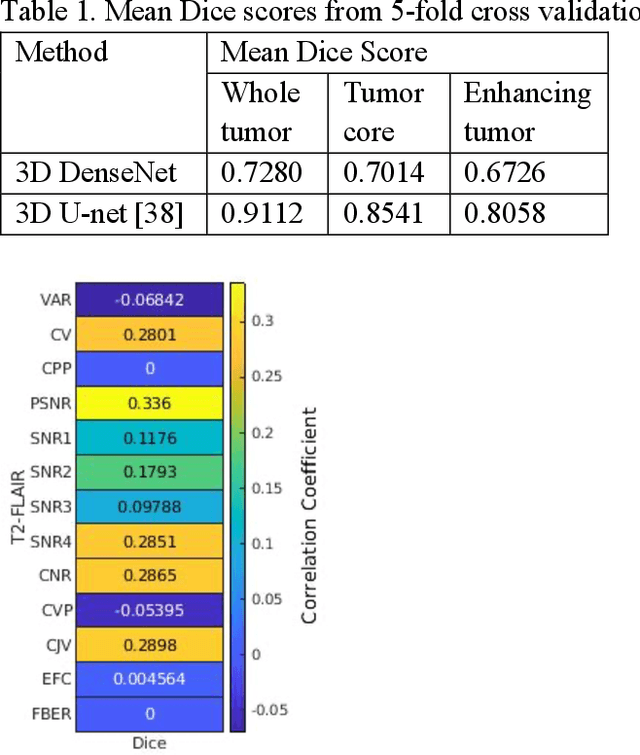
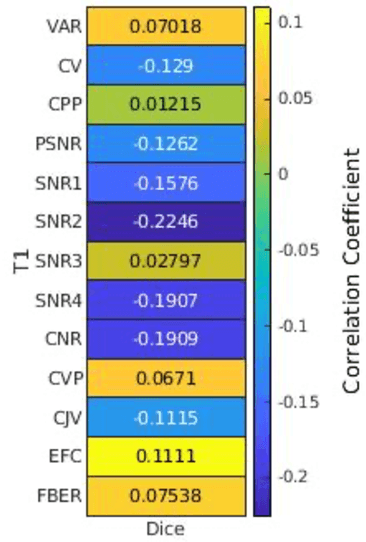
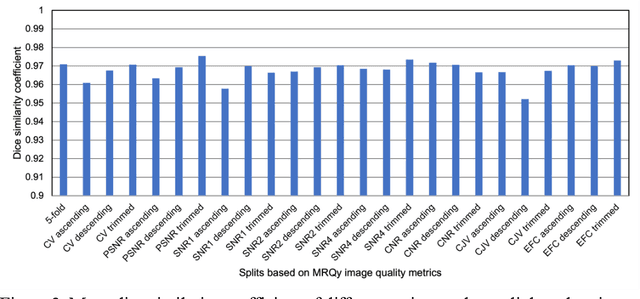
Abstract:Deep neural networks with multilevel connections process input data in complex ways to learn the information.A networks learning efficiency depends not only on the complex neural network architecture but also on the input training images.Medical image segmentation with deep neural networks for skull stripping or tumor segmentation from magnetic resonance images enables learning both global and local features of the images.Though medical images are collected in a controlled environment,there may be artifacts or equipment based variance that cause inherent bias in the input set.In this study, we investigated the correlation between the image quality metrics of MR images with the neural network segmentation accuracy.For that we have used the 3D DenseNet architecture and let the network trained on the same input but applying different methodologies to select the training data set based on the IQM values.The difference in the segmentation accuracy between models based on the random training inputs with IQM based training inputs shed light on the role of image quality metrics on segmentation accuracy.By running the image quality metrics to choose the training inputs,further we may tune the learning efficiency of the network and the segmentation accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge