Sankaran Iyer
YOLO11-4K: An Efficient Architecture for Real-Time Small Object Detection in 4K Panoramic Images
Dec 18, 2025Abstract:The processing of omnidirectional 360-degree images poses significant challenges for object detection due to inherent spatial distortions, wide fields of view, and ultra-high-resolution inputs. Conventional detectors such as YOLO are optimised for standard image sizes (for example, 640x640 pixels) and often struggle with the computational demands of 4K or higher-resolution imagery typical of 360-degree vision. To address these limitations, we introduce YOLO11-4K, an efficient real-time detection framework tailored for 4K panoramic images. The architecture incorporates a novel multi-scale detection head with a P2 layer to improve sensitivity to small objects often missed at coarser scales, and a GhostConv-based backbone to reduce computational complexity without sacrificing representational power. To enable evaluation, we manually annotated the CVIP360 dataset, generating 6,876 frame-level bounding boxes and producing a publicly available, detection-ready benchmark for 4K panoramic scenes. YOLO11-4K achieves 0.95 mAP at 0.50 IoU with 28.3 milliseconds inference per frame, representing a 75 percent latency reduction compared to YOLO11 (112.3 milliseconds), while also improving accuracy (mAP at 0.50 of 0.95 versus 0.908). This balance of efficiency and precision enables robust object detection in expansive 360-degree environments, making the framework suitable for real-world high-resolution panoramic applications. While this work focuses on 4K omnidirectional images, the approach is broadly applicable to high-resolution detection tasks in autonomous navigation, surveillance, and augmented reality.
Attention and Pooling based Sigmoid Colon Segmentation in 3D CT images
Sep 25, 2023



Abstract:Segmentation of the sigmoid colon is a crucial aspect of treating diverticulitis. It enables accurate identification and localisation of inflammation, which in turn helps healthcare professionals make informed decisions about the most appropriate treatment options. This research presents a novel deep learning architecture for segmenting the sigmoid colon from Computed Tomography (CT) images using a modified 3D U-Net architecture. Several variations of the 3D U-Net model with modified hyper-parameters were examined in this study. Pyramid pooling (PyP) and channel-spatial Squeeze and Excitation (csSE) were also used to improve the model performance. The networks were trained using manually annotated sigmoid colon. A five-fold cross-validation procedure was used on a test dataset to evaluate the network's performance. As indicated by the maximum Dice similarity coefficient (DSC) of 56.92+/-1.42%, the application of PyP and csSE techniques improves segmentation precision. We explored ensemble methods including averaging, weighted averaging, majority voting, and max ensemble. The results show that average and majority voting approaches with a threshold value of 0.5 and consistent weight distribution among the top three models produced comparable and optimal results with DSC of 88.11+/-3.52%. The results indicate that the application of a modified 3D U-Net architecture is effective for segmenting the sigmoid colon in Computed Tomography (CT) images. In addition, the study highlights the potential benefits of integrating ensemble methods to improve segmentation precision.
Organ localisation using supervised and semi supervised approaches combining reinforcement learning with imitation learning
Dec 06, 2021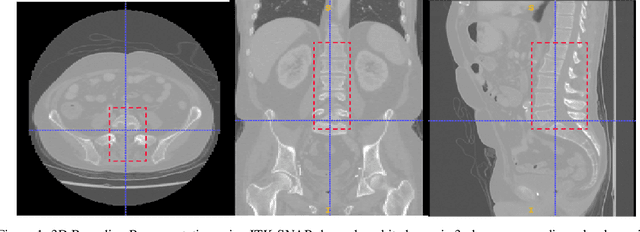
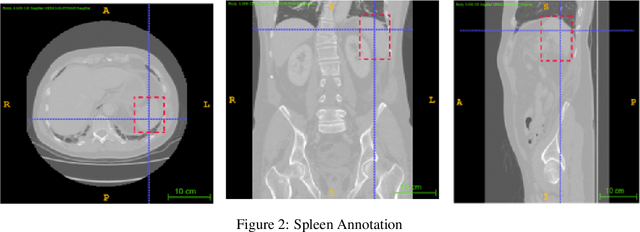
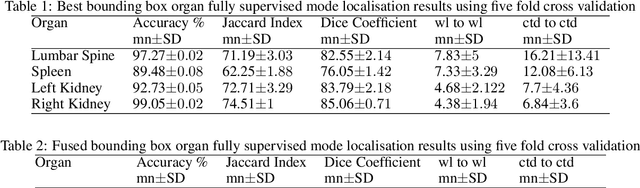
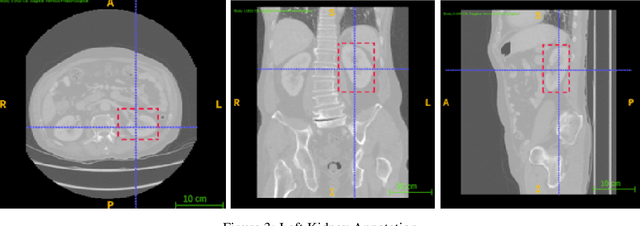
Abstract:Computer aided diagnostics often requires analysis of a region of interest (ROI) within a radiology scan, and the ROI may be an organ or a suborgan. Although deep learning algorithms have the ability to outperform other methods, they rely on the availability of a large amount of annotated data. Motivated by the need to address this limitation, an approach to localisation and detection of multiple organs based on supervised and semi-supervised learning is presented here. It draws upon previous work by the authors on localising the thoracic and lumbar spine region in CT images. The method generates six bounding boxes of organs of interest, which are then fused to a single bounding box. The results of experiments on localisation of the Spleen, Left and Right Kidneys in CT Images using supervised and semi supervised learning (SSL) demonstrate the ability to address data limitations with a much smaller data set and fewer annotations, compared to other state-of-the-art methods. The SSL performance was evaluated using three different mixes of labelled and unlabelled data (i.e.30:70,35:65,40:60) for each of lumbar spine, spleen left and right kidneys respectively. The results indicate that SSL provides a workable alternative especially in medical imaging where it is difficult to obtain annotated data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge