Daniel Moses
LWT-ARTERY-LABEL: A Lightweight Framework for Automated Coronary Artery Identification
Aug 09, 2025Abstract:Coronary artery disease (CAD) remains the leading cause of death globally, with computed tomography coronary angiography (CTCA) serving as a key diagnostic tool. However, coronary arterial analysis using CTCA, such as identifying artery-specific features from computational modelling, is labour-intensive and time-consuming. Automated anatomical labelling of coronary arteries offers a potential solution, yet the inherent anatomical variability of coronary trees presents a significant challenge. Traditional knowledge-based labelling methods fall short in leveraging data-driven insights, while recent deep-learning approaches often demand substantial computational resources and overlook critical clinical knowledge. To address these limitations, we propose a lightweight method that integrates anatomical knowledge with rule-based topology constraints for effective coronary artery labelling. Our approach achieves state-of-the-art performance on benchmark datasets, providing a promising alternative for automated coronary artery labelling.
Organ localisation using supervised and semi supervised approaches combining reinforcement learning with imitation learning
Dec 06, 2021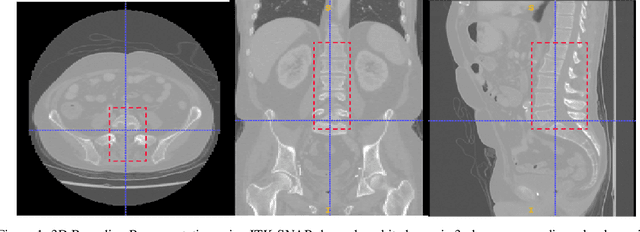
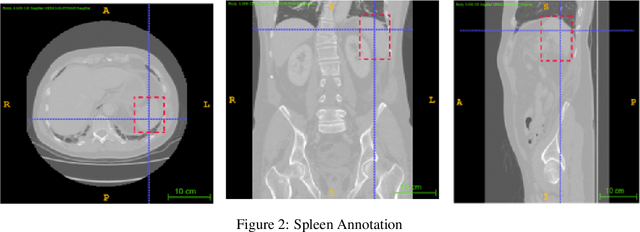
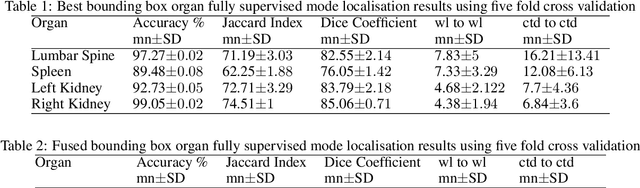
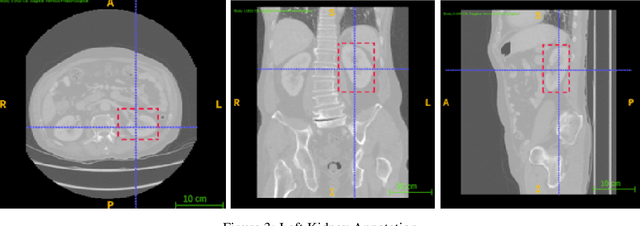
Abstract:Computer aided diagnostics often requires analysis of a region of interest (ROI) within a radiology scan, and the ROI may be an organ or a suborgan. Although deep learning algorithms have the ability to outperform other methods, they rely on the availability of a large amount of annotated data. Motivated by the need to address this limitation, an approach to localisation and detection of multiple organs based on supervised and semi-supervised learning is presented here. It draws upon previous work by the authors on localising the thoracic and lumbar spine region in CT images. The method generates six bounding boxes of organs of interest, which are then fused to a single bounding box. The results of experiments on localisation of the Spleen, Left and Right Kidneys in CT Images using supervised and semi supervised learning (SSL) demonstrate the ability to address data limitations with a much smaller data set and fewer annotations, compared to other state-of-the-art methods. The SSL performance was evaluated using three different mixes of labelled and unlabelled data (i.e.30:70,35:65,40:60) for each of lumbar spine, spleen left and right kidneys respectively. The results indicate that SSL provides a workable alternative especially in medical imaging where it is difficult to obtain annotated data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge