Sanjay Aneja
Analysis of the MICCAI Brain Tumor Segmentation -- Metastases (BraTS-METS) 2025 Lighthouse Challenge: Brain Metastasis Segmentation on Pre- and Post-treatment MRI
Apr 16, 2025Abstract:Despite continuous advancements in cancer treatment, brain metastatic disease remains a significant complication of primary cancer and is associated with an unfavorable prognosis. One approach for improving diagnosis, management, and outcomes is to implement algorithms based on artificial intelligence for the automated segmentation of both pre- and post-treatment MRI brain images. Such algorithms rely on volumetric criteria for lesion identification and treatment response assessment, which are still not available in clinical practice. Therefore, it is critical to establish tools for rapid volumetric segmentations methods that can be translated to clinical practice and that are trained on high quality annotated data. The BraTS-METS 2025 Lighthouse Challenge aims to address this critical need by establishing inter-rater and intra-rater variability in dataset annotation by generating high quality annotated datasets from four individual instances of segmentation by neuroradiologists while being recorded on video (two instances doing "from scratch" and two instances after AI pre-segmentation). This high-quality annotated dataset will be used for testing phase in 2025 Lighthouse challenge and will be publicly released at the completion of the challenge. The 2025 Lighthouse challenge will also release the 2023 and 2024 segmented datasets that were annotated using an established pipeline of pre-segmentation, student annotation, two neuroradiologists checking, and one neuroradiologist finalizing the process. It builds upon its previous edition by including post-treatment cases in the dataset. Using these high-quality annotated datasets, the 2025 Lighthouse challenge plans to test benchmark algorithms for automated segmentation of pre-and post-treatment brain metastases (BM), trained on diverse and multi-institutional datasets of MRI images obtained from patients with brain metastases.
The Brain Tumor Segmentation (BraTS-METS) Challenge 2023: Brain Metastasis Segmentation on Pre-treatment MRI
Jun 01, 2023



Abstract:Clinical monitoring of metastatic disease to the brain can be a laborious and time-consuming process, especially in cases involving multiple metastases when the assessment is performed manually. The Response Assessment in Neuro-Oncology Brain Metastases (RANO-BM) guideline, which utilizes the unidimensional longest diameter, is commonly used in clinical and research settings to evaluate response to therapy in patients with brain metastases. However, accurate volumetric assessment of the lesion and surrounding peri-lesional edema holds significant importance in clinical decision-making and can greatly enhance outcome prediction. The unique challenge in performing segmentations of brain metastases lies in their common occurrence as small lesions. Detection and segmentation of lesions that are smaller than 10 mm in size has not demonstrated high accuracy in prior publications. The brain metastases challenge sets itself apart from previously conducted MICCAI challenges on glioma segmentation due to the significant variability in lesion size. Unlike gliomas, which tend to be larger on presentation scans, brain metastases exhibit a wide range of sizes and tend to include small lesions. We hope that the BraTS-METS dataset and challenge will advance the field of automated brain metastasis detection and segmentation.
CUTS: A Fully Unsupervised Framework for Medical Image Segmentation
Sep 23, 2022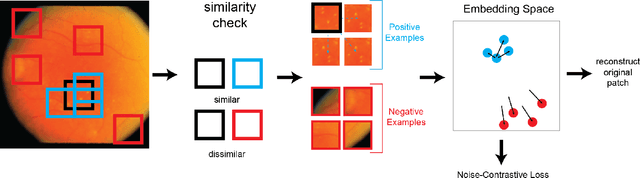
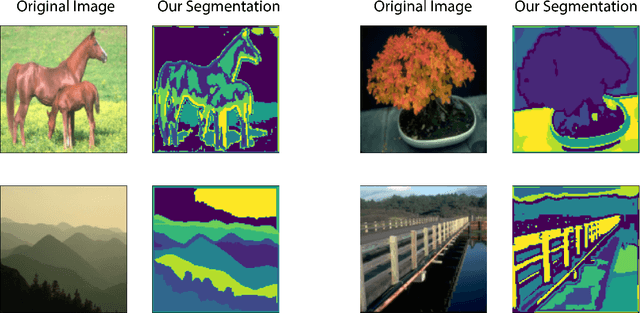
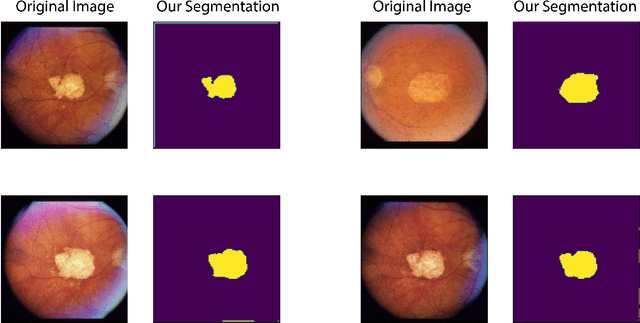
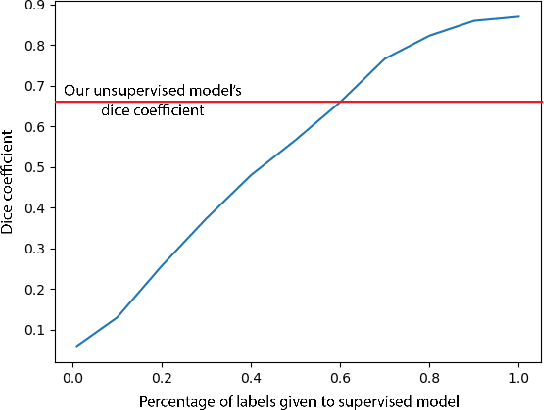
Abstract:In this work we introduce CUTS (Contrastive and Unsupervised Training for Segmentation) the first fully unsupervised deep learning framework for medical image segmentation, facilitating the use of the vast majority of imaging data that is not labeled or annotated. Segmenting medical images into regions of interest is a critical task for facilitating both patient diagnoses and quantitative research. A major limiting factor in this segmentation is the lack of labeled data, as getting expert annotations for each new set of imaging data or task can be expensive, labor intensive, and inconsistent across annotators: thus, we utilize self-supervision based on pixel-centered patches from the images themselves. Our unsupervised approach is based on a training objective with both contrastive learning and autoencoding aspects. Previous contrastive learning approaches for medical image segmentation have focused on image-level contrastive training, rather than our intra-image patch-level approach or have used this as a pre-training task where the network needed further supervised training afterwards. By contrast, we build the first entirely unsupervised framework that operates at the pixel-centered-patch level. Specifically, we add novel augmentations, a patch reconstruction loss, and introduce a new pixel clustering and identification framework. Our model achieves improved results on several key medical imaging tasks, as verified by held-out expert annotations on the task of segmenting geographic atrophy (GA) regions of images of the retina.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge