Ron O. Dror
Enhancing Ligand Pose Sampling for Molecular Docking
Nov 30, 2023Abstract:Deep learning promises to dramatically improve scoring functions for molecular docking, leading to substantial advances in binding pose prediction and virtual screening. To train scoring functions-and to perform molecular docking-one must generate a set of candidate ligand binding poses. Unfortunately, the sampling protocols currently used to generate candidate poses frequently fail to produce any poses close to the correct, experimentally determined pose, unless information about the correct pose is provided. This limits the accuracy of learned scoring functions and molecular docking. Here, we describe two improved protocols for pose sampling: GLOW (auGmented sampLing with sOftened vdW potential) and a novel technique named IVES (IteratiVe Ensemble Sampling). Our benchmarking results demonstrate the effectiveness of our methods in improving the likelihood of sampling accurate poses, especially for binding pockets whose shape changes substantially when different ligands bind. This improvement is observed across both experimentally determined and AlphaFold-generated protein structures. Additionally, we present datasets of candidate ligand poses generated using our methods for each of around 5,000 protein-ligand cross-docking pairs, for training and testing scoring functions. To benefit the research community, we provide these cross-docking datasets and an open-source Python implementation of GLOW and IVES at https://github.com/drorlab/GLOW_IVES .
FlexVDW: A machine learning approach to account for protein flexibility in ligand docking
Mar 20, 2023Abstract:Most widely used ligand docking methods assume a rigid protein structure. This leads to problems when the structure of the target protein deforms upon ligand binding. In particular, the ligand's true binding pose is often scored very unfavorably due to apparent clashes between ligand and protein atoms, which lead to extremely high values of the calculated van der Waals energy term. Traditionally, this problem has been addressed by explicitly searching for receptor conformations to account for the flexibility of the receptor in ligand binding. Here we present a deep learning model trained to take receptor flexibility into account implicitly when predicting van der Waals energy. We show that incorporating this machine-learned energy term into a state-of-the-art physics-based scoring function improves small molecule ligand pose prediction results in cases with substantial protein deformation, without degrading performance in cases with minimal protein deformation. This work demonstrates the feasibility of learning effects of protein flexibility on ligand binding without explicitly modeling changes in protein structure.
Equivariant Graph Neural Networks for 3D Macromolecular Structure
Jun 07, 2021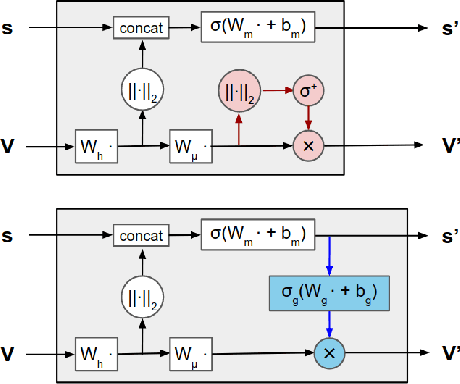
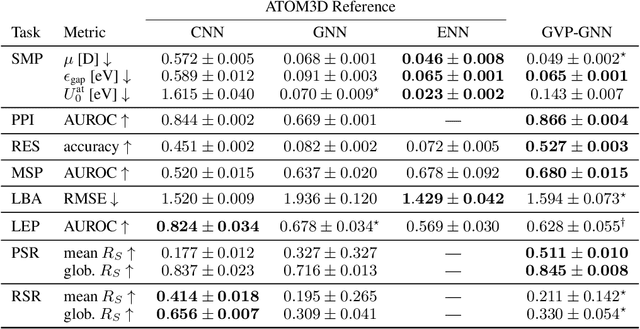
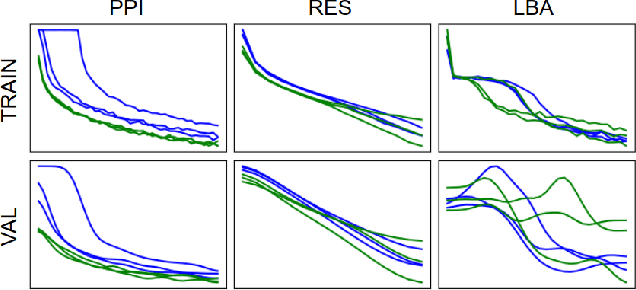
Abstract:Representing and reasoning about 3D structures of macromolecules is emerging as a distinct challenge in machine learning. Here, we extend recent work on geometric vector perceptrons and apply equivariant graph neural networks to a wide range of tasks from structural biology. Our method outperforms all reference architectures on 4 out of 8 tasks in the ATOM3D benchmark and broadly improves over rotation-invariant graph neural networks. We also demonstrate that transfer learning can improve performance in learning from macromolecular structure.
ATOM3D: Tasks On Molecules in Three Dimensions
Dec 07, 2020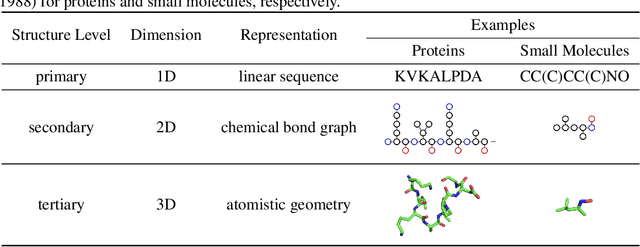
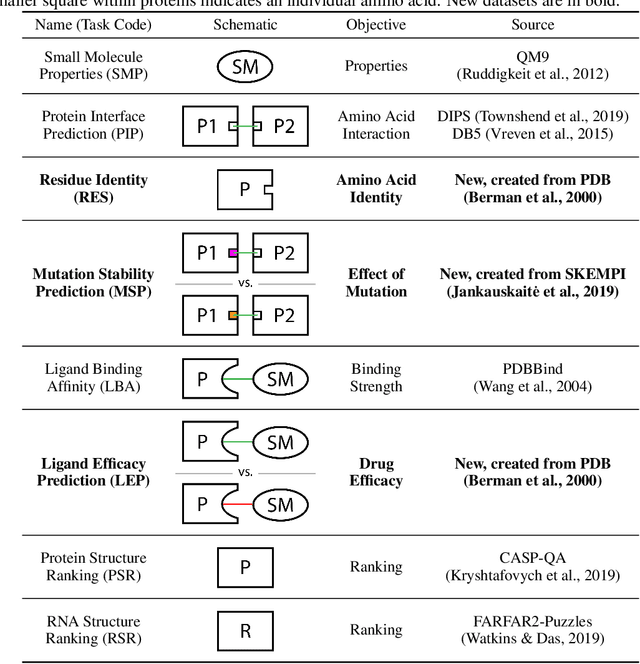
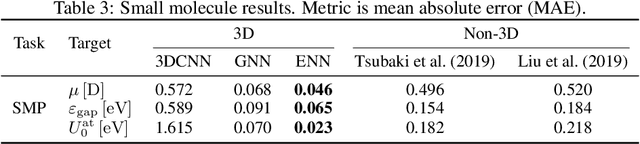
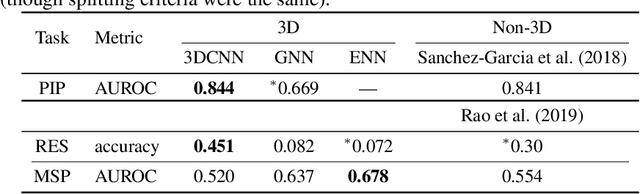
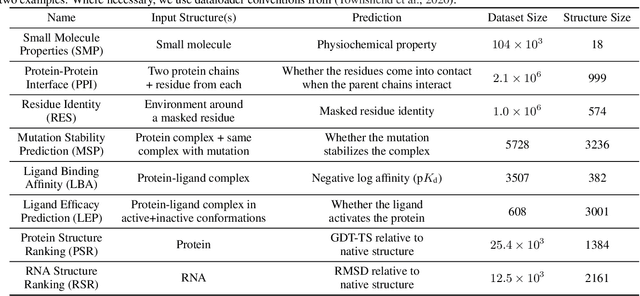
Abstract:Computational methods that operate directly on three-dimensional molecular structure hold large potential to solve important questions in biology and chemistry. In particular deep neural networks have recently gained significant attention. In this work we present ATOM3D, a collection of both novel and existing datasets spanning several key classes of biomolecules, to systematically assess such learning methods. We develop three-dimensional molecular learning networks for each of these tasks, finding that they consistently improve performance relative to one- and two-dimensional methods. The specific choice of architecture proves to be critical for performance, with three-dimensional convolutional networks excelling at tasks involving complex geometries, while graph networks perform well on systems requiring detailed positional information. Furthermore, equivariant networks show significant promise. Our results indicate many molecular problems stand to gain from three-dimensional molecular learning. All code and datasets can be accessed via https://www.atom3d.ai .
Protein model quality assessment using rotation-equivariant, hierarchical neural networks
Nov 27, 2020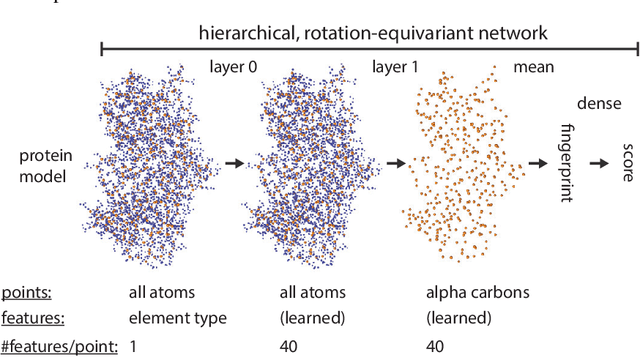
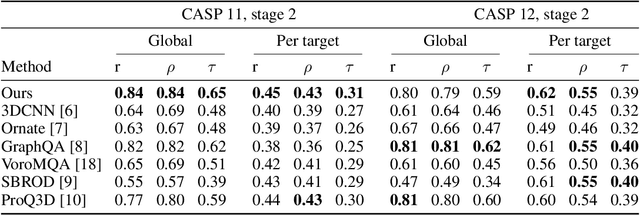
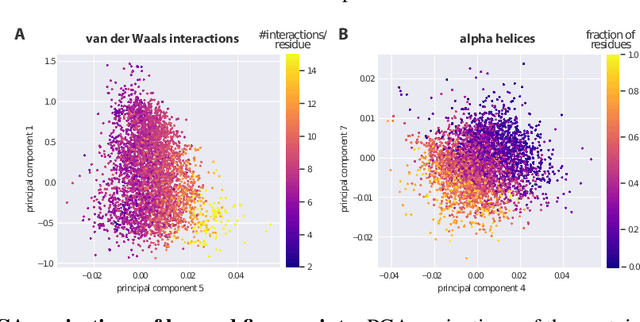
Abstract:Proteins are miniature machines whose function depends on their three-dimensional (3D) structure. Determining this structure computationally remains an unsolved grand challenge. A major bottleneck involves selecting the most accurate structural model among a large pool of candidates, a task addressed in model quality assessment. Here, we present a novel deep learning approach to assess the quality of a protein model. Our network builds on a point-based representation of the atomic structure and rotation-equivariant convolutions at different levels of structural resolution. These combined aspects allow the network to learn end-to-end from entire protein structures. Our method achieves state-of-the-art results in scoring protein models submitted to recent rounds of CASP, a blind prediction community experiment. Particularly striking is that our method does not use physics-inspired energy terms and does not rely on the availability of additional information (beyond the atomic structure of the individual protein model), such as sequence alignments of multiple proteins.
Geometric Prediction: Moving Beyond Scalars
Jun 25, 2020



Abstract:Many quantities we are interested in predicting are geometric tensors; we refer to this class of problems as geometric prediction. Attempts to perform geometric prediction in real-world scenarios have been limited to approximating them through scalar predictions, leading to losses in data efficiency. In this work, we demonstrate that equivariant networks have the capability to predict real-world geometric tensors without the need for such approximations. We show the applicability of this method to the prediction of force fields and then propose a novel formulation of an important task, biomolecular structure refinement, as a geometric prediction problem, improving state-of-the-art structural candidates. In both settings, we find that our equivariant network is able to generalize to unseen systems, despite having been trained on small sets of examples. This novel and data-efficient ability to predict real-world geometric tensors opens the door to addressing many problems through the lens of geometric prediction, in areas such as 3D vision, robotics, and molecular and structural biology.
Generalizable Protein Interface Prediction with End-to-End Learning
Sep 21, 2018

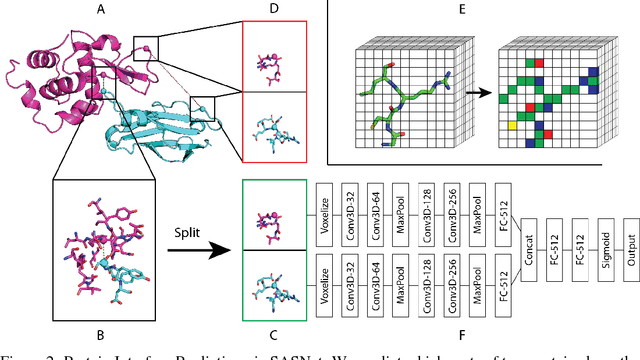
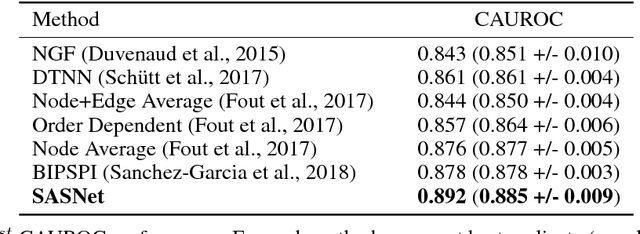
Abstract:Predicting how proteins interact with one another - that is, which surfaces of one protein bind to which surfaces of another protein - is a central problem in biology. Here we present Siamese Atomic Surfacelet Network (SASNet), the first end-to-end learning method for protein interface prediction. Despite using only spatial coordinates and identities of atoms as inputs, SASNet outperforms state-of-the-art methods that rely on complex, hand-selected features. These results are particularly striking because we train the method entirely on a significantly biased data set that does not account for the fact that proteins deform when binding to one another. Nonetheless, our network maintains high performance, without retraining, when tested on real cases in which proteins do deform. This suggests that it has learned fundamental properties of protein structure and dynamics, which has important implications for a variety of key problems related to biomolecular structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge