Qiran Jia
A False Discovery Rate Control Method Using a Fully Connected Hidden Markov Random Field for Neuroimaging Data
May 29, 2025Abstract:False discovery rate (FDR) control methods are essential for voxel-wise multiple testing in neuroimaging data analysis, where hundreds of thousands or even millions of tests are conducted to detect brain regions associated with disease-related changes. Classical FDR control methods (e.g., BH, q-value, and LocalFDR) assume independence among tests and often lead to high false non-discovery rates (FNR). Although various spatial FDR control methods have been developed to improve power, they still fall short of jointly addressing three major challenges in neuroimaging applications: capturing complex spatial dependencies, maintaining low variability in both false discovery proportion (FDP) and false non-discovery proportion (FNP) across replications, and achieving computational scalability for high-resolution data. To address these challenges, we propose fcHMRF-LIS, a powerful, stable, and scalable spatial FDR control method for voxel-wise multiple testing. It integrates the local index of significance (LIS)-based testing procedure with a novel fully connected hidden Markov random field (fcHMRF) designed to model complex spatial structures using a parsimonious parameterization. We develop an efficient expectation-maximization algorithm incorporating mean-field approximation, the Conditional Random Fields as Recurrent Neural Networks (CRF-RNN) technique, and permutohedral lattice filtering, reducing the time complexity from quadratic to linear in the number of tests. Extensive simulations demonstrate that fcHMRF-LIS achieves accurate FDR control, lower FNR, reduced variability in FDP and FNP, and a higher number of true positives compared to existing methods. Applied to an FDG-PET dataset from the Alzheimer's Disease Neuroimaging Initiative, fcHMRF-LIS identifies neurobiologically relevant brain regions and offers notable advantages in computational efficiency.
DeepFDR: A Deep Learning-based False Discovery Rate Control Method for Neuroimaging Data
Oct 20, 2023



Abstract:Voxel-based multiple testing is widely used in neuroimaging data analysis. Traditional false discovery rate (FDR) control methods often ignore the spatial dependence among the voxel-based tests and thus suffer from substantial loss of testing power. While recent spatial FDR control methods have emerged, their validity and optimality remain questionable when handling the complex spatial dependencies of the brain. Concurrently, deep learning methods have revolutionized image segmentation, a task closely related to voxel-based multiple testing. In this paper, we propose DeepFDR, a novel spatial FDR control method that leverages unsupervised deep learning-based image segmentation to address the voxel-based multiple testing problem. Numerical studies, including comprehensive simulations and Alzheimer's disease FDG-PET image analysis, demonstrate DeepFDR's superiority over existing methods. DeepFDR not only excels in FDR control and effectively diminishes the false nondiscovery rate, but also boasts exceptional computational efficiency highly suited for tackling large-scale neuroimaging data.
BiTr-Unet: a CNN-Transformer Combined Network for MRI Brain Tumor Segmentation
Sep 25, 2021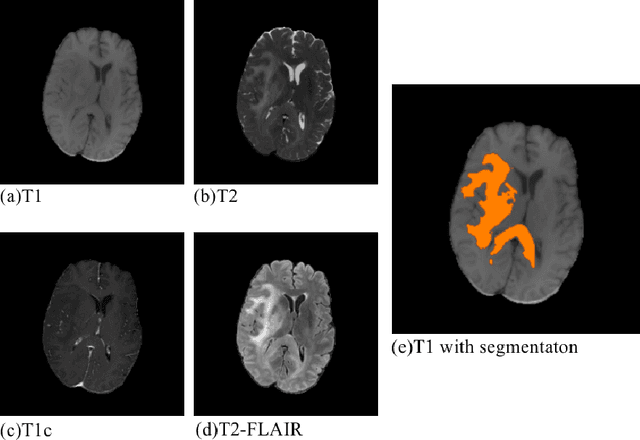
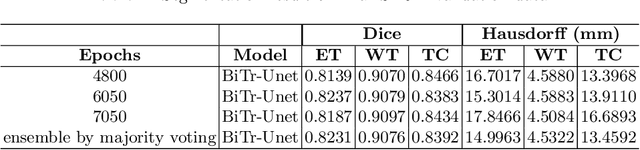
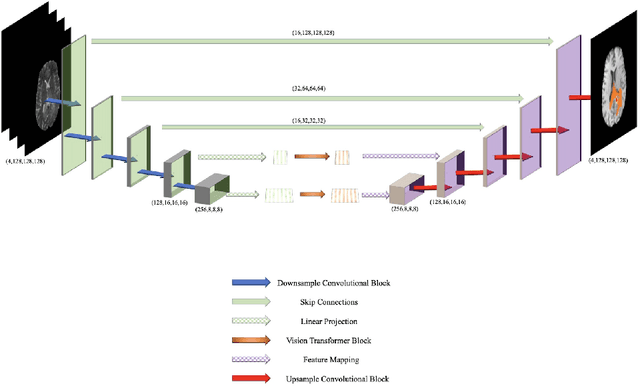
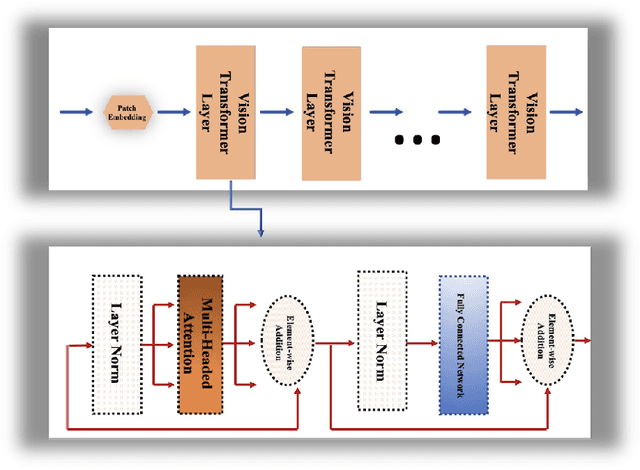
Abstract:Convolutional neural networks (CNNs) have recently achieved remarkable success in automatically identifying organs or lesions on 3D medical images. Meanwhile, vision transformer networks have exhibited exceptional performance in 2D image classification tasks. Compared with CNNs, transformer networks have an obvious advantage of extracting long-range features due to their self-attention algorithm. Therefore, in this paper we present a CNN-Transformer combined model called BiTr-Unet for brain tumor segmentation on multi-modal MRI scans. The proposed BiTr-Unet achieves good performance on the BraTS 2021 validation dataset with mean Dice score 0.9076, 0.8392 and 0.8231, and mean Hausdorff distance 4.5322, 13.4592 and 14.9963 for the whole tumor, tumor core, and enhancing tumor, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge