Prashanti Manda
Semantic Analysis of SNOMED CT Concept Co-occurrences in Clinical Documentation using MIMIC-IV
Sep 03, 2025Abstract:Clinical notes contain rich clinical narratives but their unstructured format poses challenges for large-scale analysis. Standardized terminologies such as SNOMED CT improve interoperability, yet understanding how concepts relate through co-occurrence and semantic similarity remains underexplored. In this study, we leverage the MIMIC-IV database to investigate the relationship between SNOMED CT concept co-occurrence patterns and embedding-based semantic similarity. Using Normalized Pointwise Mutual Information (NPMI) and pretrained embeddings (e.g., ClinicalBERT, BioBERT), we examine whether frequently co-occurring concepts are also semantically close, whether embeddings can suggest missing concepts, and how these relationships evolve temporally and across specialties. Our analyses reveal that while co-occurrence and semantic similarity are weakly correlated, embeddings capture clinically meaningful associations not always reflected in documentation frequency. Embedding-based suggestions frequently matched concepts later documented, supporting their utility for augmenting clinical annotations. Clustering of concept embeddings yielded coherent clinical themes (symptoms, labs, diagnoses, cardiovascular conditions) that map to patient phenotypes and care patterns. Finally, co-occurrence patterns linked to outcomes such as mortality and readmission demonstrate the practical utility of this approach. Collectively, our findings highlight the complementary value of co-occurrence statistics and semantic embeddings in improving documentation completeness, uncovering latent clinical relationships, and informing decision support and phenotyping applications.
Information-theoretic Interestingness Measures for Cross-Ontology Data Mining
May 16, 2016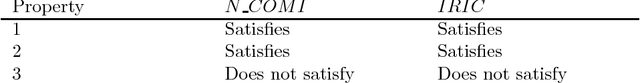
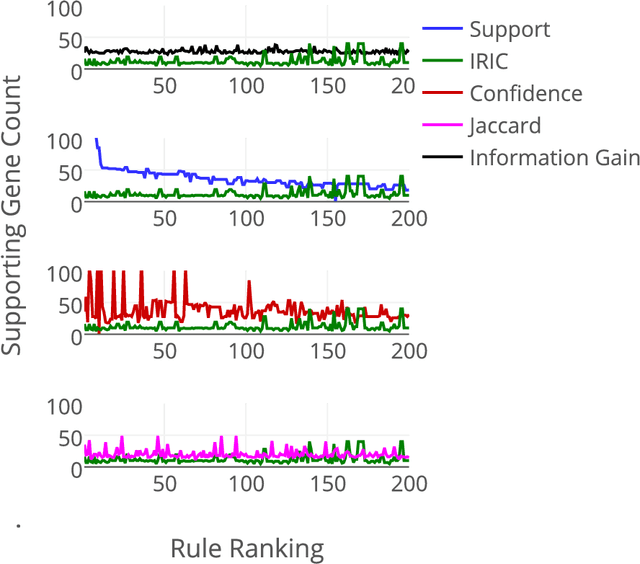
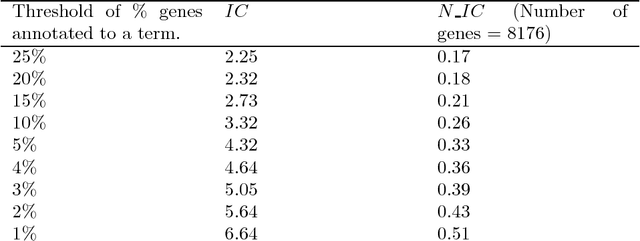
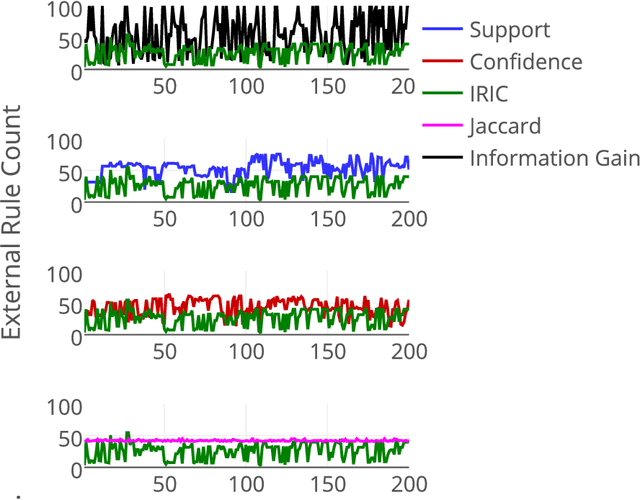
Abstract:Community annotation of biological entities with concepts from multiple bio-ontologies has created large and growing repositories of ontology-based annotation data with embedded implicit relationships among orthogonal ontologies. Development of efficient data mining methods and metrics to mine and assess the quality of the mined relationships has not kept pace with the growth of annotation data. In this study, we present a data mining method that uses ontology-guided generalization to discover relationships across ontologies along with a new interestingness metric based on information theory. We apply our data mining algorithm and interestingness measures to datasets from the Gene Expression Database at the Mouse Genome Informatics as a preliminary proof of concept to mine relationships between developmental stages in the mouse anatomy ontology and Gene Ontology concepts (biological process, molecular function and cellular component). In addition, we present a comparison of our interestingness metric to four existing metrics. Ontology-based annotation datasets provide a valuable resource for discovery of relationships across ontologies. The use of efficient data mining methods and appropriate interestingness metrics enables the identification of high quality relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge