Pierrick Coupe
An Explainable Diagnostic Framework for Neurodegenerative Dementias via Reinforcement-Optimized LLM Reasoning
May 26, 2025Abstract:The differential diagnosis of neurodegenerative dementias is a challenging clinical task, mainly because of the overlap in symptom presentation and the similarity of patterns observed in structural neuroimaging. To improve diagnostic efficiency and accuracy, deep learning-based methods such as Convolutional Neural Networks and Vision Transformers have been proposed for the automatic classification of brain MRIs. However, despite their strong predictive performance, these models find limited clinical utility due to their opaque decision making. In this work, we propose a framework that integrates two core components to enhance diagnostic transparency. First, we introduce a modular pipeline for converting 3D T1-weighted brain MRIs into textual radiology reports. Second, we explore the potential of modern Large Language Models (LLMs) to assist clinicians in the differential diagnosis between Frontotemporal dementia subtypes, Alzheimer's disease, and normal aging based on the generated reports. To bridge the gap between predictive accuracy and explainability, we employ reinforcement learning to incentivize diagnostic reasoning in LLMs. Without requiring supervised reasoning traces or distillation from larger models, our approach enables the emergence of structured diagnostic rationales grounded in neuroimaging findings. Unlike post-hoc explainability methods that retrospectively justify model decisions, our framework generates diagnostic rationales as part of the inference process-producing causally grounded explanations that inform and guide the model's decision-making process. In doing so, our framework matches the diagnostic performance of existing deep learning methods while offering rationales that support its diagnostic conclusions.
vol2Brain: A new online Pipeline for whole Brain MRI analysis
Feb 08, 2022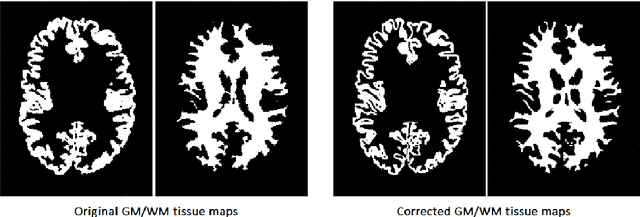
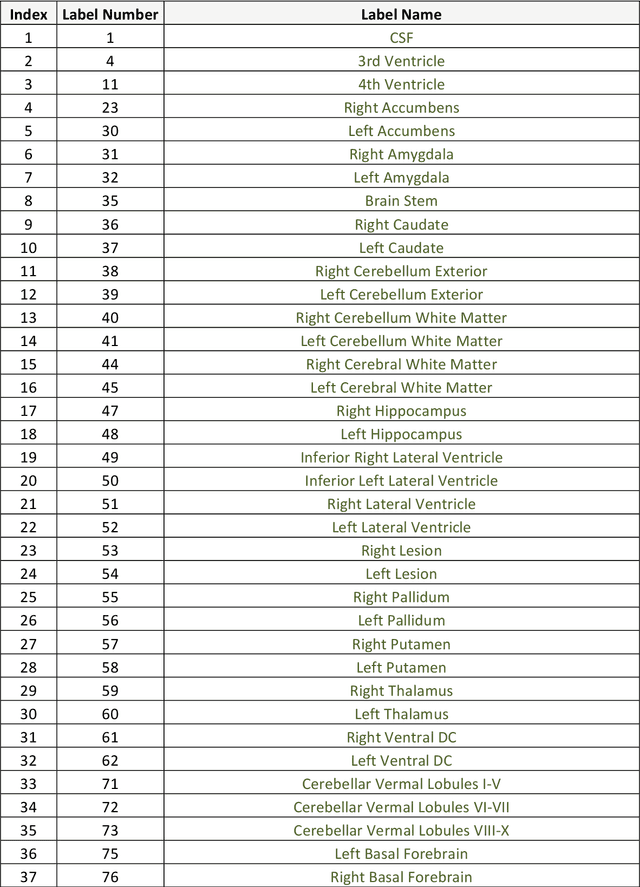
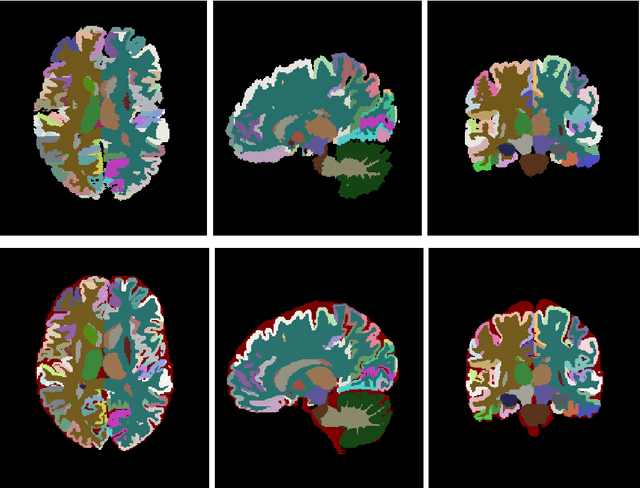
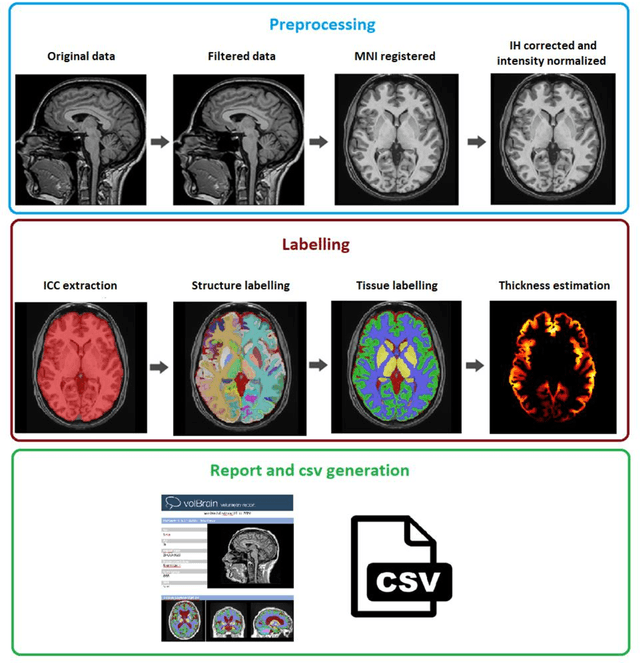
Abstract:Automatic and reliable quantitative tools for MR brain image analysis are a very valuable resources for both clinical and research environments. In the last years, this field has experienced many advances with successful techniques based on label fusion and more recently deep learning. However, few of them have been specifically designed to provide a dense anatomical labelling at multiscale level and to deal with brain anatomical alterations such as white matter lesions. In this work, we present a fully automatic pipeline (vol2Brain) for whole brain segmentation and analysis which densely labels (N>100) the brain while being robust to the presence of white matter lesions. This new pipeline is an evolution of our previous volBrain pipeline that extends significantly the number of regions that can be analyzed. Our proposed method is based on a fast multiscale multi-atlas label fusion technology with systematic error correction able to provide accurate volumetric information in few minutes. We have deployed our new pipeline within our platform volBrain (www.volbrain.upv.es) which has been already demonstrated to be an efficient and effective manner to share our technology with users worldwide
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge