Noriaki Sato
DAG Learning from Zero-Inflated Count Data Using Continuous Optimization
Dec 18, 2025



Abstract:We address network structure learning from zero-inflated count data by casting each node as a zero-inflated generalized linear model and optimizing a smooth, score-based objective under a directed acyclic graph constraint. Our Zero-Inflated Continuous Optimization (ZICO) approach uses node-wise likelihoods with canonical links and enforces acyclicity through a differentiable surrogate constraint combined with sparsity regularization. ZICO achieves superior performance with faster runtimes on simulated data. It also performs comparably to or better than common algorithms for reverse engineering gene regulatory networks. ZICO is fully vectorized and mini-batched, enabling learning on larger variable sets with practical runtimes in a wide range of domains.
Practical Causal Evaluation Metrics for Biological Networks
Nov 16, 2025Abstract:Estimating causal networks from biological data is a critical step in systems biology. When evaluating the inferred network, assessing the networks based on their intervention effects is particularly important for downstream probabilistic reasoning and the identification of potential drug targets. In the context of gene regulatory network inference, biological databases are often used as reference sources. These databases typically describe relationships in a qualitative rather than quantitative manner. However, few evaluation metrics have been developed that take this qualitative nature into account. To address this, we developed a metric, the sign-augmented Structural Intervention Distance (sSID), and a weighted sSID that incorporates the net effects of the intervention. Through simulations and analyses of real transcriptomic datasets, we found that our proposed metrics could identify a different algorithm as optimal compared to conventional metrics, and the network selected by sSID had a superior performance in the classification task of clinical covariates using transcriptomic data. This suggests that sSID can distinguish networks that are structurally correct but functionally incorrect, highlighting its potential as a more biologically meaningful and practical evaluation metric.
Individual health-disease phase diagrams for disease prevention based on machine learning
May 31, 2022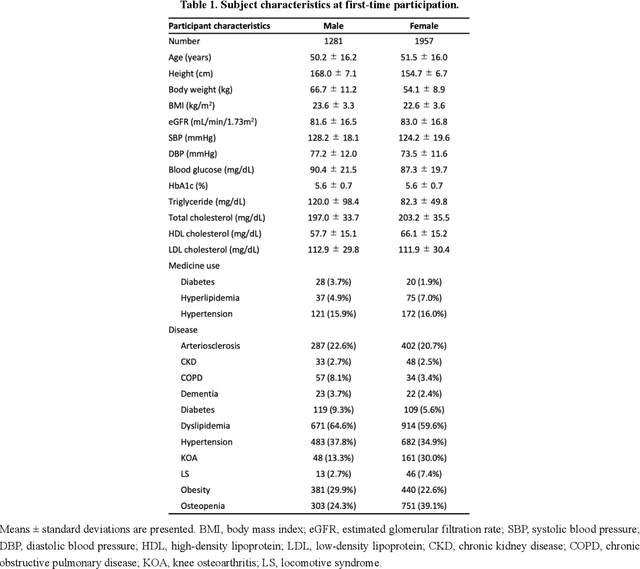
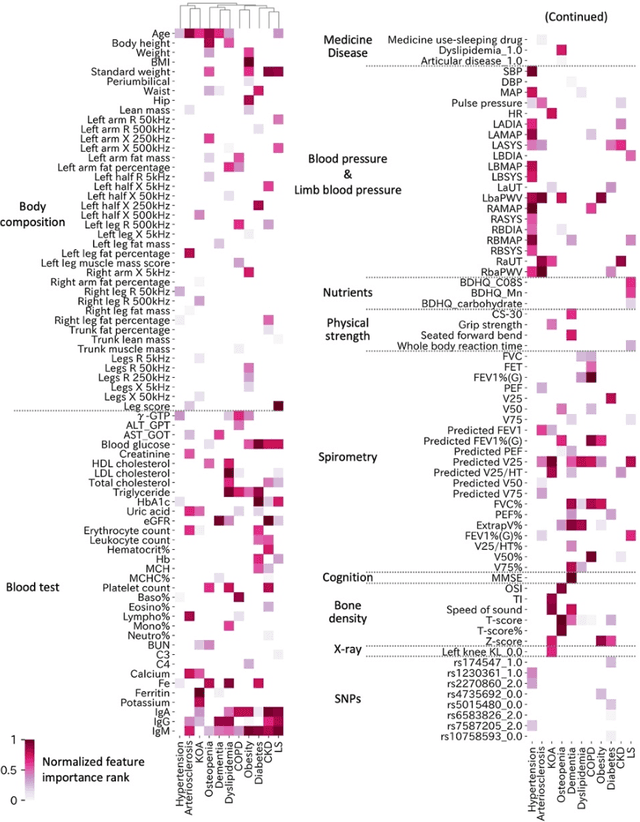
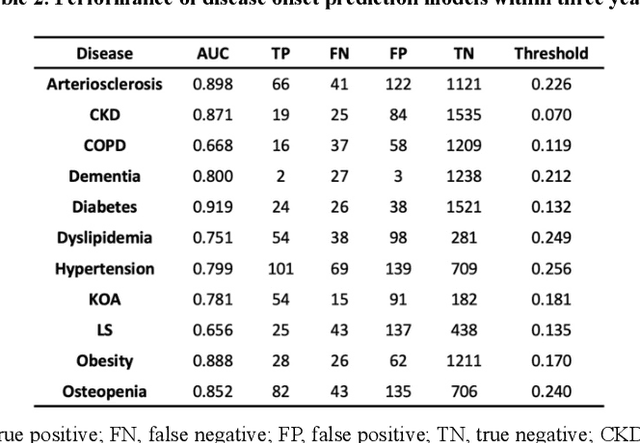
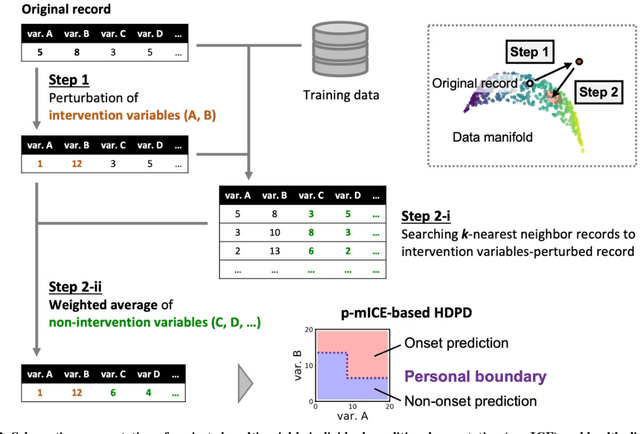
Abstract:Early disease detection and prevention methods based on effective interventions are gaining attention. Machine learning technology has enabled precise disease prediction by capturing individual differences in multivariate data. Progress in precision medicine has revealed that substantial heterogeneity exists in health data at the individual level and that complex health factors are involved in the development of chronic diseases. However, it remains a challenge to identify individual physiological state changes in cross-disease onset processes because of the complex relationships among multiple biomarkers. Here, we present the health-disease phase diagram (HDPD), which represents a personal health state by visualizing the boundary values of multiple biomarkers that fluctuate early in the disease progression process. In HDPDs, future onset predictions are represented by perturbing multiple biomarker values while accounting for dependencies among variables. We constructed HDPDs for 11 non-communicable diseases (NCDs) from a longitudinal health checkup cohort of 3,238 individuals, comprising 3,215 measurement items and genetic data. Improvement of biomarker values to the non-onset region in HDPD significantly prevented future disease onset in 7 out of 11 NCDs. Our results demonstrate that HDPDs can represent individual physiological states in the onset process and be used as intervention goals for disease prevention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge