Naoki Nonaka
Syn-STARTS: Synthesized START Triage Scenario Generation Framework for Scalable LLM Evaluation
Nov 18, 2025



Abstract:Triage is a critically important decision-making process in mass casualty incidents (MCIs) to maximize victim survival rates. While the role of AI in such situations is gaining attention for making optimal decisions within limited resources and time, its development and performance evaluation require benchmark datasets of sufficient quantity and quality. However, MCIs occur infrequently, and sufficient records are difficult to accumulate at the scene, making it challenging to collect large-scale realworld data for research use. Therefore, we developed Syn-STARTS, a framework that uses LLMs to generate triage cases, and verified its effectiveness. The results showed that the triage cases generated by Syn-STARTS were qualitatively indistinguishable from the TRIAGE open dataset generated by manual curation from training materials. Furthermore, when evaluating the LLM accuracy using hundreds of cases each from the green, yellow, red, and black categories defined by the standard triage method START, the results were found to be highly stable. This strongly indicates the possibility of synthetic data in developing high-performance AI models for severe and critical medical situations.
Efficient HLA imputation from sequential SNPs data by Transformer
Nov 11, 2022Abstract:Human leukocyte antigen (HLA) genes are associated with a variety of diseases, however direct typing of HLA is time and cost consuming. Thus various imputation methods using sequential SNPs data have been proposed based on statistical or deep learning models, e.g. CNN-based model, named DEEP*HLA. However, imputation efficiency is not sufficient for in frequent alleles and a large size of reference panel is required. Here, we developed a Transformer-based model to impute HLA alleles, named "HLA Reliable IMputatioN by Transformer (HLARIMNT)" to take advantage of sequential nature of SNPs data. We validated the performance of HLARIMNT using two different reference panels; Pan-Asian reference panel (n = 530) and Type 1 Diabetes Genetics Consortium (T1DGC) reference panel (n = 5,225), as well as the mixture of those two panels (n = 1,060). HLARIMNT achieved higher accuracy than DEEP*HLA by several indices, especially for infrequent alleles. We also varied the size of data used for training, and HLARIMNT imputed more accurately among any size of training data. These results suggest that Transformer-based model may impute efficiently not only HLA types but also any other gene types from sequential SNPs data.
General-to-Detailed GAN for Infrequent Class Medical Images
Nov 28, 2018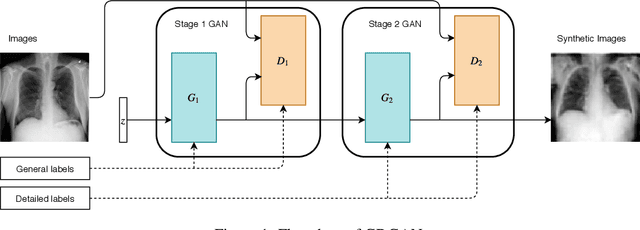

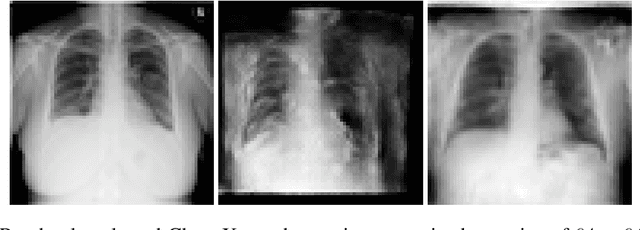
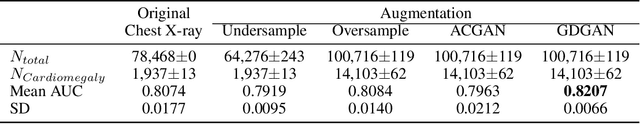
Abstract:Deep learning has significant potential for medical imaging. However, since the incident rate of each disease varies widely, the frequency of classes in a medical image dataset is imbalanced, leading to poor accuracy for such infrequent classes. One possible solution is data augmentation of infrequent classes using synthesized images created by Generative Adversarial Networks (GANs), but conventional GANs also require certain amount of images to learn. To overcome this limitation, here we propose General-to-detailed GAN (GDGAN), serially connected two GANs, one for general labels and the other for detailed labels. GDGAN produced diverse medical images, and the network trained with an augmented dataset outperformed other networks using existing methods with respect to Area-Under-Curve (AUC) of Receiver Operating Characteristic (ROC) curve.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge