Morteza Babaie
Kimia Lab, University of Waterloo, Waterloo, ON, Canada, Vector Institute, MaRS Centre, Toronto, Canada
A new Local Radon Descriptor for Content-Based Image Search
Jul 30, 2020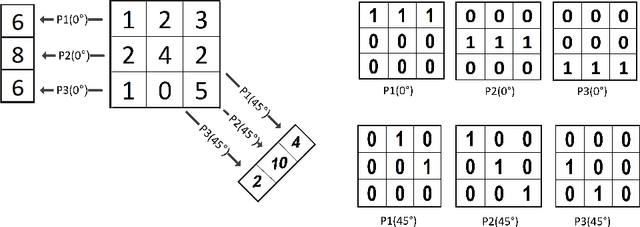
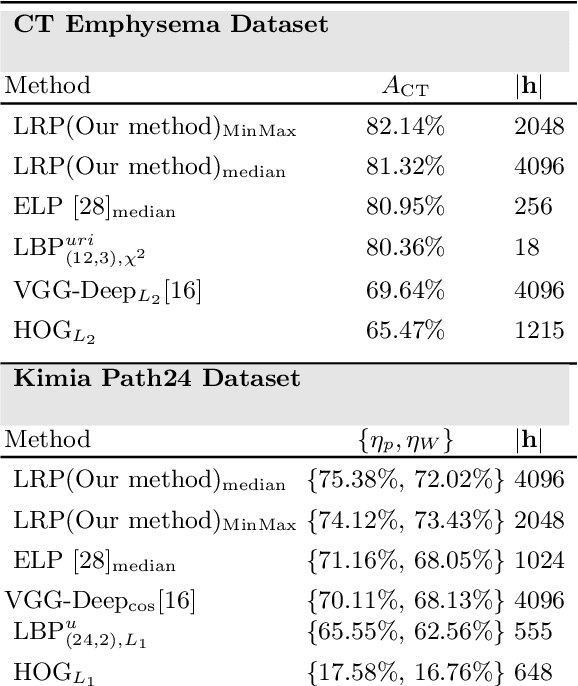
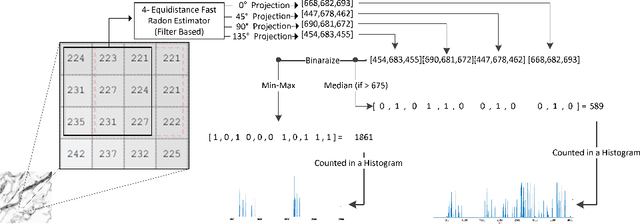
Abstract:Content-based image retrieval (CBIR) is an essential part of computer vision research, especially in medical expert systems. Having a discriminative image descriptor with the least number of parameters for tuning is desirable in CBIR systems. In this paper, we introduce a new simple descriptor based on the histogram of local Radon projections. We also propose a very fast convolution-based local Radon estimator to overcome the slow process of Radon projections. We performed our experiments using pathology images (KimiaPath24) and lung CT patches and test our proposed solution for medical image processing. We achieved superior results compared with other histogram-based descriptors such as LBP and HoG as well as some pre-trained CNNs.
Recognizing Magnification Levels in Microscopic Snapshots
May 07, 2020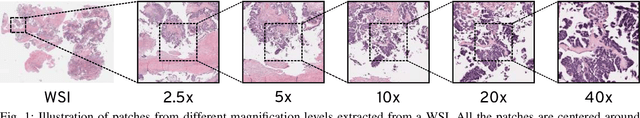
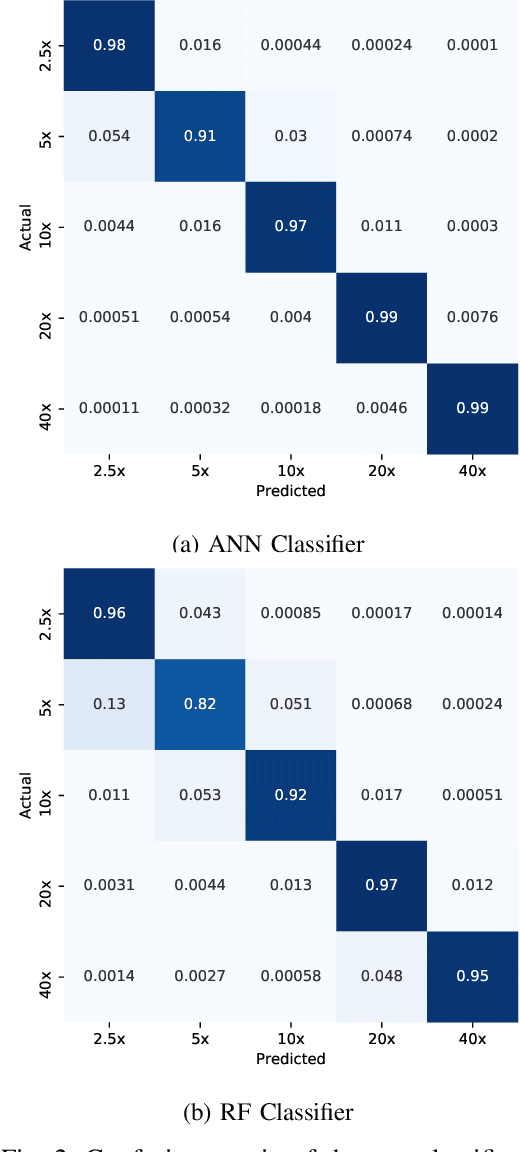
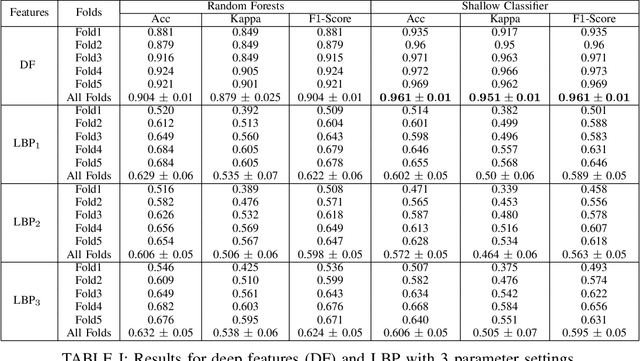
Abstract:Recent advances in digital imaging has transformed computer vision and machine learning to new tools for analyzing pathology images. This trend could automate some of the tasks in the diagnostic pathology and elevate the pathologist workload. The final step of any cancer diagnosis procedure is performed by the expert pathologist. These experts use microscopes with high level of optical magnification to observe minute characteristics of the tissue acquired through biopsy and fixed on glass slides. Switching between different magnifications, and finding the magnification level at which they identify the presence or absence of malignant tissues is important. As the majority of pathologists still use light microscopy, compared to digital scanners, in many instance a mounted camera on the microscope is used to capture snapshots from significant field-of-views. Repositories of such snapshots usually do not contain the magnification information. In this paper, we extract deep features of the images available on TCGA dataset with known magnification to train a classifier for magnification recognition. We compared the results with LBP, a well-known handcrafted feature extraction method. The proposed approach achieved a mean accuracy of 96% when a multi-layer perceptron was trained as a classifier.
Pan-Cancer Diagnostic Consensus Through Searching Archival Histopathology Images Using Artificial Intelligence
Nov 20, 2019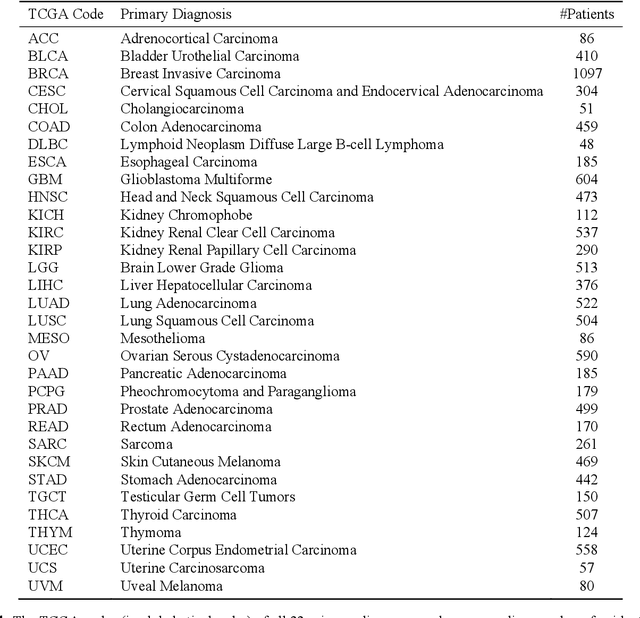
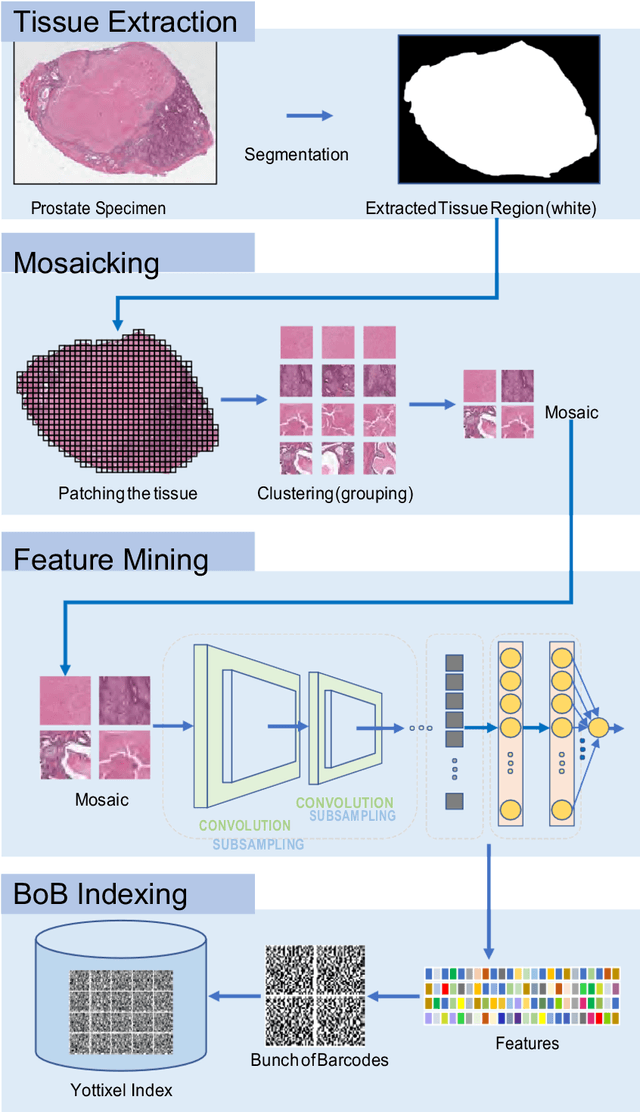
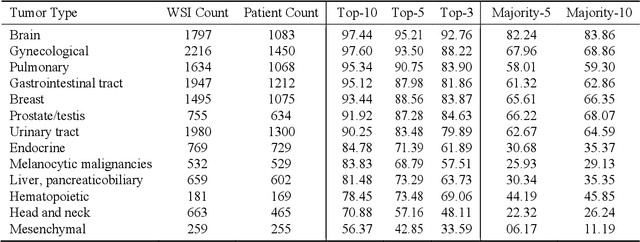
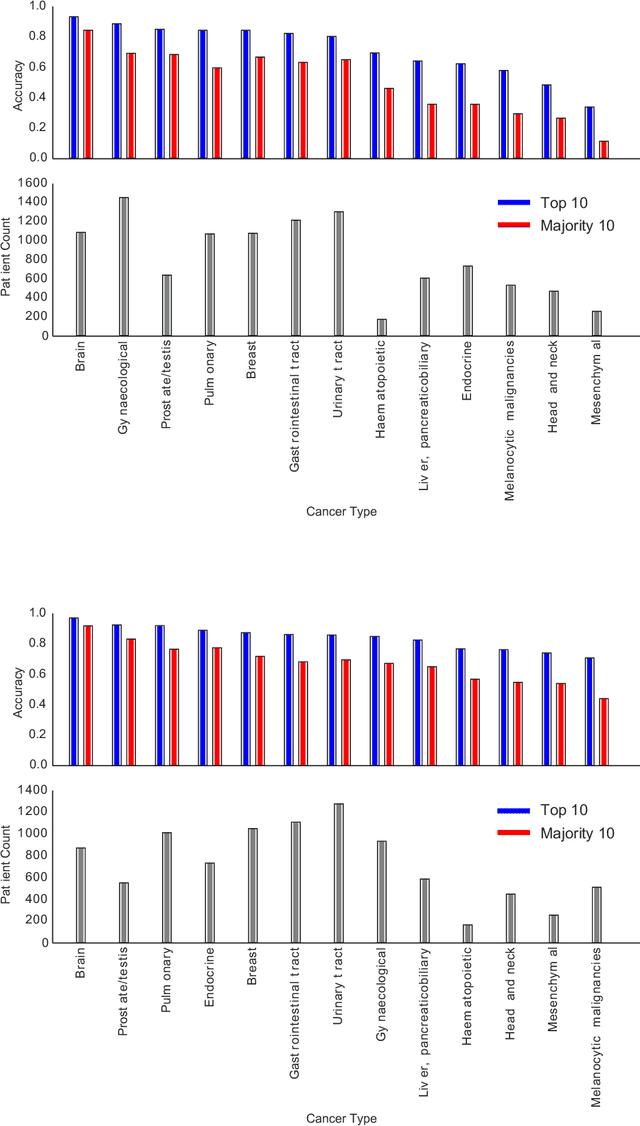
Abstract:The emergence of digital pathology has opened new horizons for histopathology and cytology. Artificial-intelligence algorithms are able to operate on digitized slides to assist pathologists with diagnostic tasks. Whereas machine learning involving classification and segmentation methods have obvious benefits for image analysis in pathology, image search represents a fundamental shift in computational pathology. Matching the pathology of new patients with already diagnosed and curated cases offers pathologist a novel approach to improve diagnostic accuracy through visual inspection of similar cases and computational majority vote for consensus building. In this study, we report the results from searching the largest public repository (The Cancer Genome Atlas [TCGA] program by National Cancer Institute, USA) of whole slide images from almost 11,000 patients depicting different types of malignancies. For the first time, we successfully indexed and searched almost 30,000 high-resolution digitized slides constituting 16 terabytes of data comprised of 20 million 1000x1000 pixels image patches. The TCGA image database covers 25 anatomic sites and contains 32 cancer subtypes. High-performance storage and GPU power were employed for experimentation. The results were assessed with conservative "majority voting" to build consensus for subtype diagnosis through vertical search and demonstrated high accuracy values for both frozen sections slides (e.g., bladder urothelial carcinoma 93%, kidney renal clear cell carcinoma 97%, and ovarian serous cystadenocarcinoma 99%) and permanent histopathology slides (e.g., prostate adenocarcinoma 98%, skin cutaneous melanoma 99%, and thymoma 100%). The key finding of this validation study was that computational consensus appears to be possible for rendering diagnoses if a sufficiently large number of searchable cases are available for each cancer subtype.
Subtractive Perceptrons for Learning Images: A Preliminary Report
Sep 15, 2019
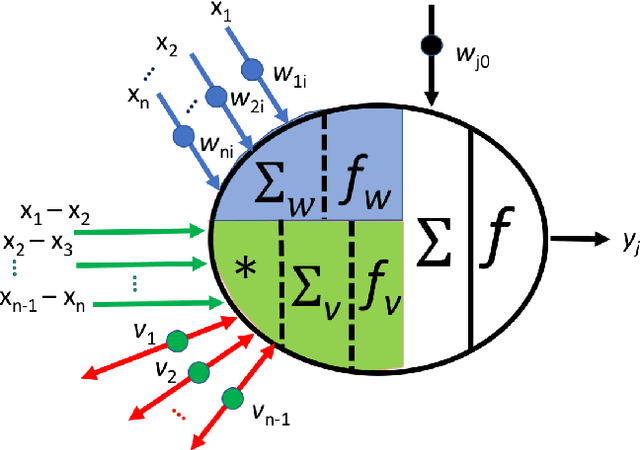
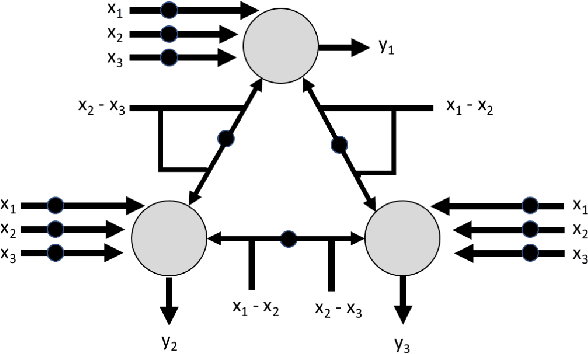
Abstract:In recent years, artificial neural networks have achieved tremendous success for many vision-based tasks. However, this success remains within the paradigm of \emph{weak AI} where networks, among others, are specialized for just one given task. The path toward \emph{strong AI}, or Artificial General Intelligence, remains rather obscure. One factor, however, is clear, namely that the feed-forward structure of current networks is not a realistic abstraction of the human brain. In this preliminary work, some ideas are proposed to define a \textit{subtractive Perceptron} (s-Perceptron), a graph-based neural network that delivers a more compact topology to learn one specific task. In this preliminary study, we test the s-Perceptron with the MNIST dataset, a commonly used image archive for digit recognition. The proposed network achieves excellent results compared to the benchmark networks that rely on more complex topologies.
Patch Clustering for Representation of Histopathology Images
Mar 17, 2019
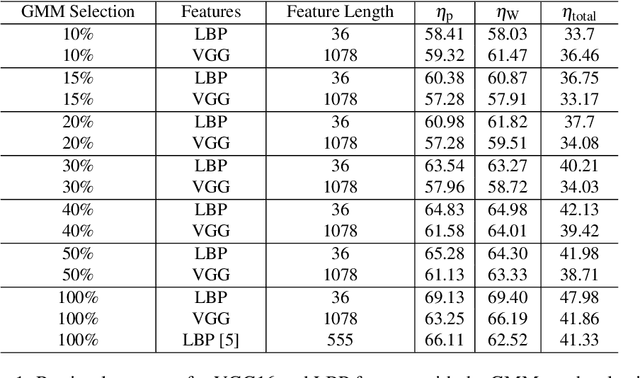
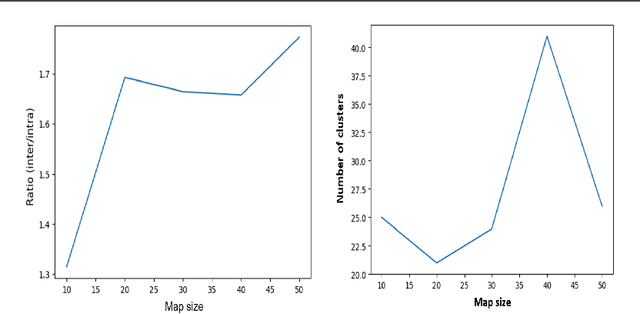
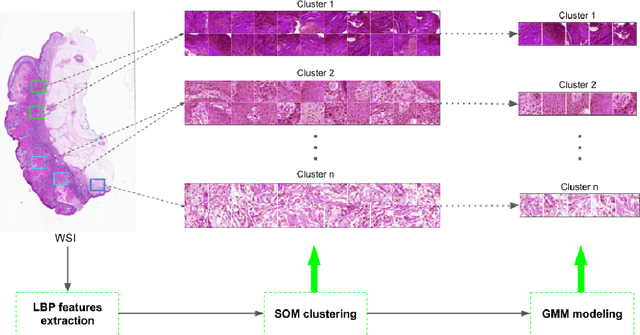
Abstract:Whole Slide Imaging (WSI) has become an important topic during the last decade. Even though significant progress in both medical image processing and computational resources has been achieved, there are still problems in WSI that need to be solved. A major challenge is the scan size. The dimensions of digitized tissue samples may exceed 100,000 by 100,000 pixels causing memory and efficiency obstacles for real-time processing. The main contribution of this work is representing a WSI by selecting a small number of patches for algorithmic processing (e.g., indexing and search). As a result, we reduced the search time and storage by various factors between ($50\% - 90\%$), while losing only a few percentages in the patch retrieval accuracy. A self-organizing map (SOM) has been applied on local binary patterns (LBP) and deep features of the KimiaPath24 dataset in order to cluster patches that share the same characteristics. We used a Gaussian mixture model (GMM) to represent each class with a rather small ($10\%-50\%$) portion of patches. The results showed that LBP features can outperform deep features. By selecting only $50\%$ of all patches after SOM clustering and GMM patch selection, we received $65\%$ accuracy for retrieval of the best match, while the maximum accuracy (using all patches) was $69\%$.
Deep Features for Tissue-Fold Detection in Histopathology Images
Mar 17, 2019
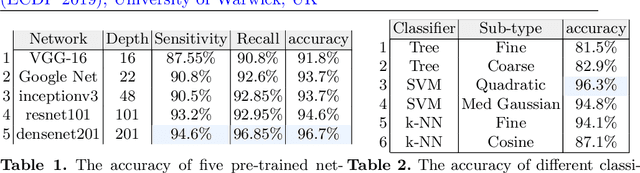
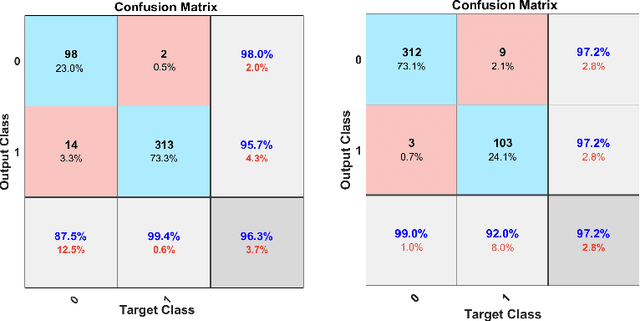

Abstract:Whole slide imaging (WSI) refers to the digitization of a tissue specimen which enables pathologists to explore high-resolution images on a monitor rather than through a microscope. The formation of tissue folds occur during tissue processing. Their presence may not only cause out-of-focus digitization but can also negatively affect the diagnosis in some cases. In this paper, we have compared five pre-trained convolutional neural networks (CNNs) of different depths as feature extractors to characterize tissue folds. We have also explored common classifiers to discriminate folded tissue against the normal tissue in hematoxylin and eosin (H\&E) stained biopsy samples. In our experiments, we manually select the folded area in roughly 2.5mm $\times$ 2.5mm patches at $20$x magnification level as the training data. The ``DenseNet'' with 201 layers alongside an SVM classifier outperformed all other configurations. Based on the leave-one-out validation strategy, we achieved $96.3\%$ accuracy, whereas with augmentation the accuracy increased to $97.2\%$. We have tested the generalization of our method with five unseen WSIs from the NIH (National Cancer Institute) dataset. The accuracy for patch-wise detection was $81\%$. One folded patch within an image suffices to flag the entire specimen for visual inspection.
Facial Recognition with Encoded Local Projections
Sep 11, 2018
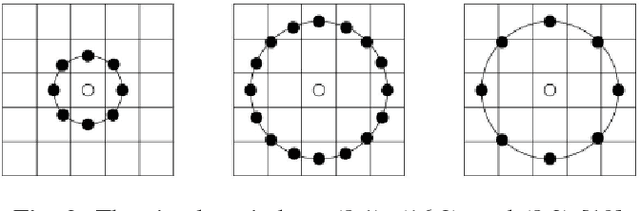

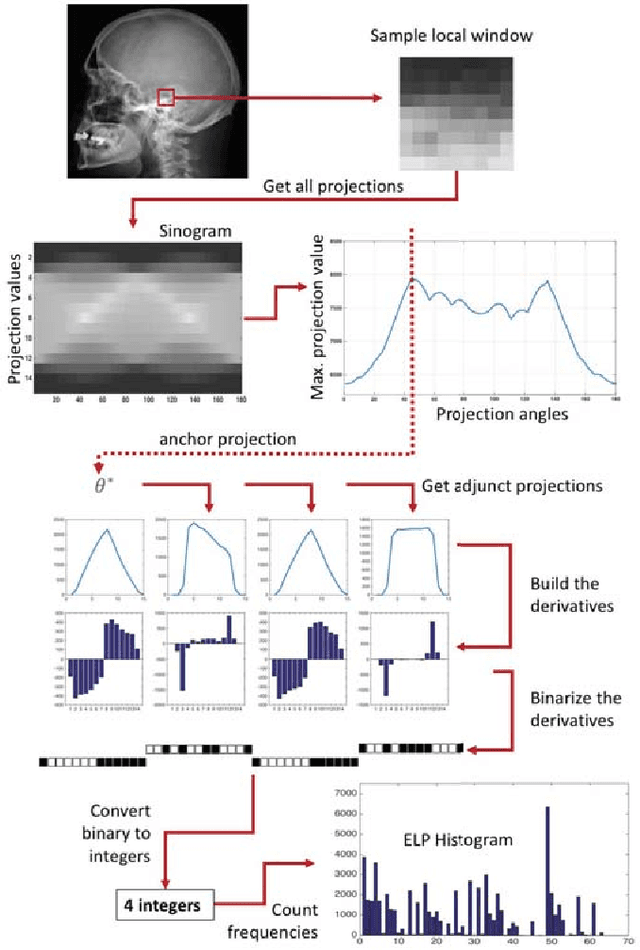
Abstract:Encoded Local Projections (ELP) is a recently introduced dense sampling image descriptor which uses projections in small neighbourhoods to construct a histogram/descriptor for the entire image. ELP has shown to be as accurate as other state-of-the-art features in searching medical images while being time and resource efficient. This paper attempts for the first time to utilize ELP descriptor as primary features for facial recognition and compare the results with LBP histogram on the Labeled Faces in the Wild dataset. We have evaluated descriptors by comparing the chi-squared distance of each image descriptor versus all others as well as training Support Vector Machines (SVM) with each feature vector. In both cases, the results of ELP were better than LBP in the same sub-image configuration.
Deep Barcodes for Fast Retrieval of Histopathology Scans
Apr 30, 2018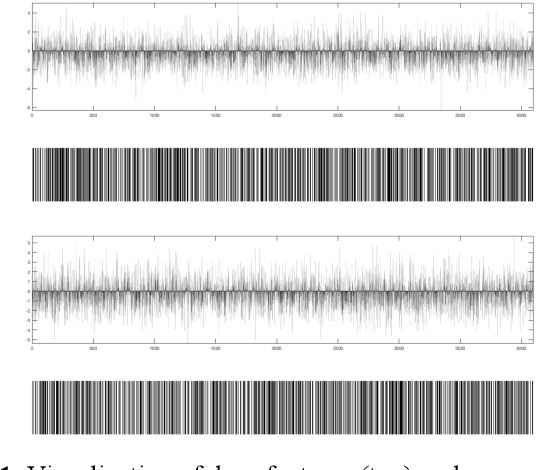
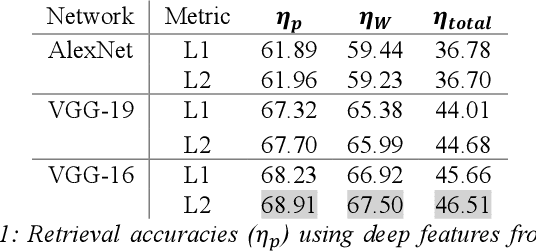
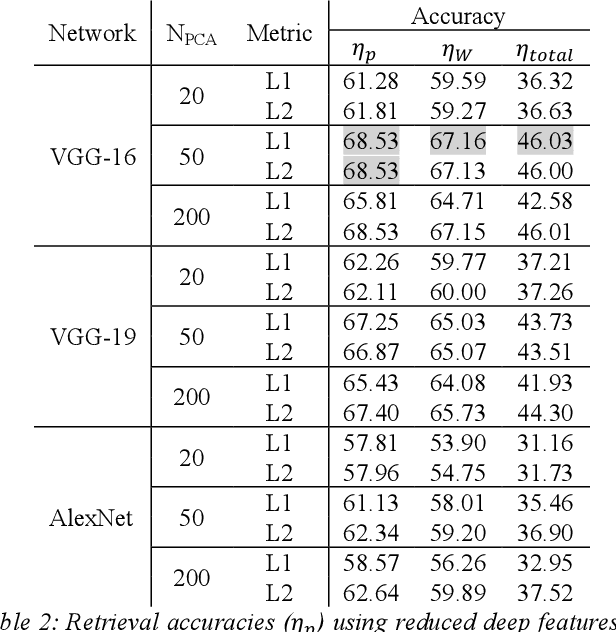
Abstract:We investigate the concept of deep barcodes and propose two methods to generate them in order to expedite the process of classification and retrieval of histopathology images. Since binary search is computationally less expensive, in terms of both speed and storage, deep barcodes could be useful when dealing with big data retrieval. Our experiments use the dataset Kimia Path24 to test three pre-trained networks for image retrieval. The dataset consists of 27,055 training images in 24 different classes with large variability, and 1,325 test images for testing. Apart from the high-speed and efficiency, results show a surprising retrieval accuracy of 71.62% for deep barcodes, as compared to 68.91% for deep features and 68.53% for compressed deep features.
Local Radon Descriptors for Image Search
Oct 11, 2017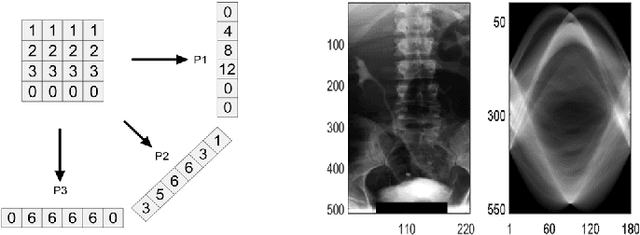
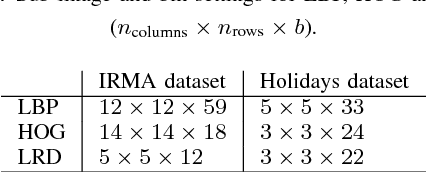
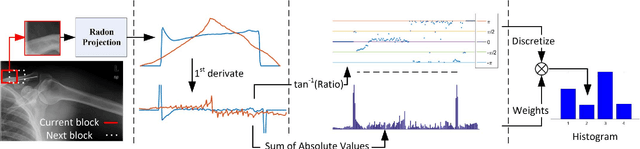
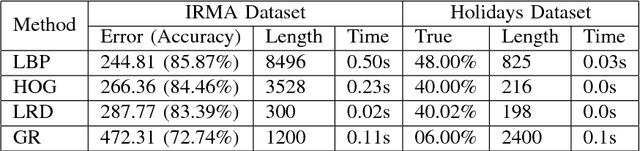
Abstract:Radon transform and its inverse operation are important techniques in medical imaging tasks. Recently, there has been renewed interest in Radon transform for applications such as content-based medical image retrieval. However, all studies so far have used Radon transform as a global or quasi-global image descriptor by extracting projections of the whole image or large sub-images. This paper attempts to show that the dense sampling to generate the histogram of local Radon projections has a much higher discrimination capability than the global one. In this paper, we introduce Local Radon Descriptor (LRD) and apply it to the IRMA dataset, which contains 14,410 x-ray images as well as to the INRIA Holidays dataset with 1,990 images. Our results show significant improvement in retrieval performance by using LRD versus its global version. We also demonstrate that LRD can deliver results comparable to well-established descriptors like LBP and HOG.
Convolutional Neural Networks for Histopathology Image Classification: Training vs. Using Pre-Trained Networks
Oct 11, 2017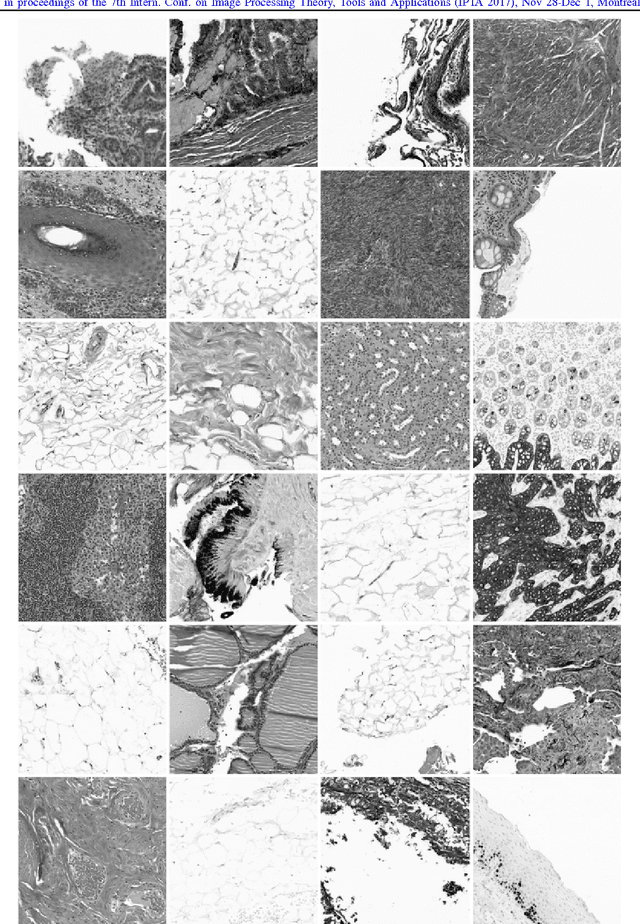

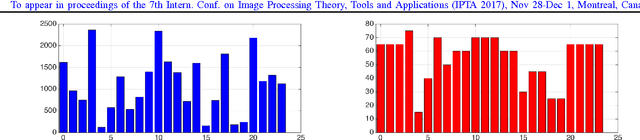
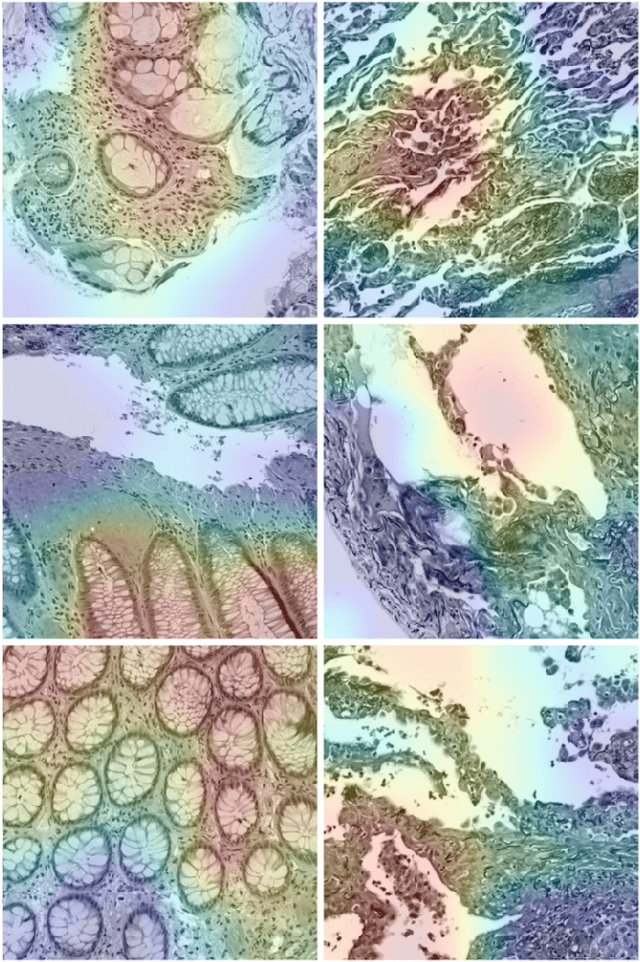
Abstract:We explore the problem of classification within a medical image data-set based on a feature vector extracted from the deepest layer of pre-trained Convolution Neural Networks. We have used feature vectors from several pre-trained structures, including networks with/without transfer learning to evaluate the performance of pre-trained deep features versus CNNs which have been trained by that specific dataset as well as the impact of transfer learning with a small number of samples. All experiments are done on Kimia Path24 dataset which consists of 27,055 histopathology training patches in 24 tissue texture classes along with 1,325 test patches for evaluation. The result shows that pre-trained networks are quite competitive against training from scratch. As well, fine-tuning does not seem to add any tangible improvement for VGG16 to justify additional training while we observed considerable improvement in retrieval and classification accuracy when we fine-tuned the Inception structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge