Mona Alshahrani
Multi-View MRI Approach for Classification of MGMT Methylation in Glioblastoma Patients
Dec 16, 2025Abstract:The presence of MGMT promoter methylation significantly affects how well chemotherapy works for patients with Glioblastoma Multiforme (GBM). Currently, confirmation of MGMT promoter methylation relies on invasive brain tumor tissue biopsies. In this study, we explore radiogenomics techniques, a promising approach in precision medicine, to identify genetic markers from medical images. Using MRI scans and deep learning models, we propose a new multi-view approach that considers spatial relationships between MRI views to detect MGMT methylation status. Importantly, our method extracts information from all three views without using a complicated 3D deep learning model, avoiding issues associated with high parameter count, slow convergence, and substantial memory demands. We also introduce a new technique for tumor slice extraction and show its superiority over existing methods based on multiple evaluation metrics. By comparing our approach to state-of-the-art models, we demonstrate the efficacy of our method. Furthermore, we share a reproducible pipeline of published models, encouraging transparency and the development of robust diagnostic tools. Our study highlights the potential of non-invasive methods for identifying MGMT promoter methylation and contributes to advancing precision medicine in GBM treatment.
Neuro-symbolic representation learning on biological knowledge graphs
Dec 13, 2016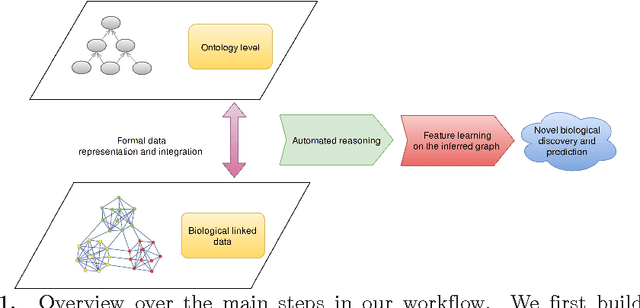
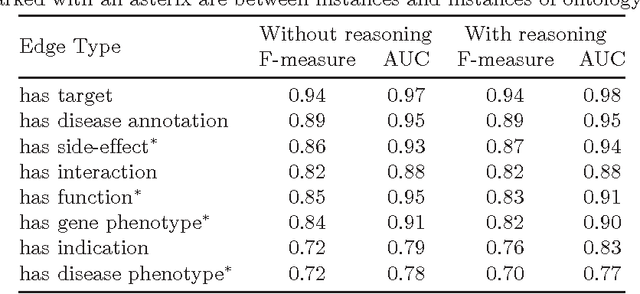
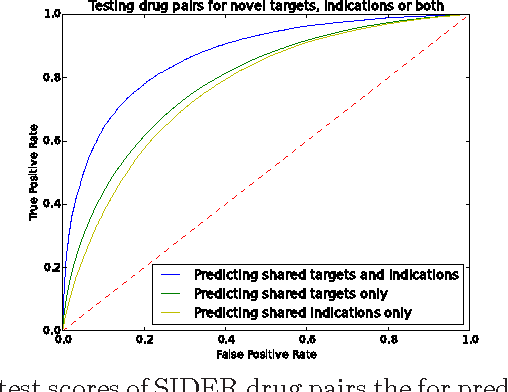
Abstract:Motivation: Biological data and knowledge bases increasingly rely on Semantic Web technologies and the use of knowledge graphs for data integration, retrieval and federated queries. In the past years, feature learning methods that are applicable to graph-structured data are becoming available, but have not yet widely been applied and evaluated on structured biological knowledge. Results: We develop a novel method for feature learning on biological knowledge graphs. Our method combines symbolic methods, in particular knowledge representation using symbolic logic and automated reasoning, with neural networks to generate embeddings of nodes that encode for related information within knowledge graphs. Through the use of symbolic logic, these embeddings contain both explicit and implicit information. We apply these embeddings to the prediction of edges in the knowledge graph representing problems of function prediction, finding candidate genes of diseases, protein-protein interactions, or drug target relations, and demonstrate performance that matches and sometimes outperforms traditional approaches based on manually crafted features. Our method can be applied to any biological knowledge graph, and will thereby open up the increasing amount of Semantic Web based knowledge bases in biology to use in machine learning and data analytics. Availability and Implementation: https://github.com/bio-ontology-research-group/walking-rdf-and-owl Contact: robert.hoehndorf@kaust.edu.sa
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge