Matthias Samwald
A critical analysis of metrics used for measuring progress in artificial intelligence
Aug 06, 2020
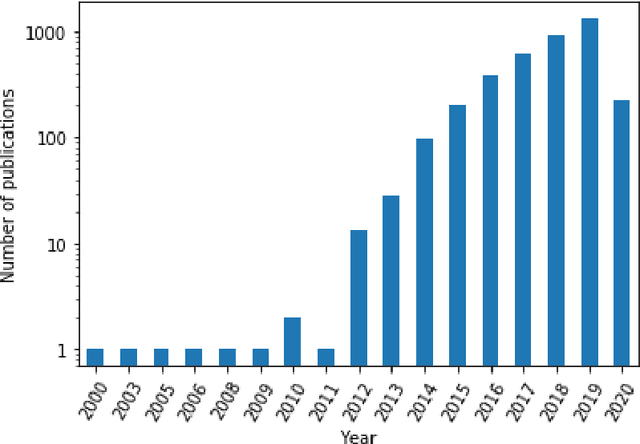
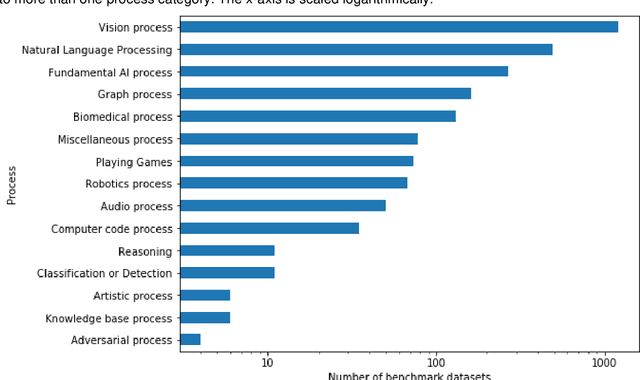
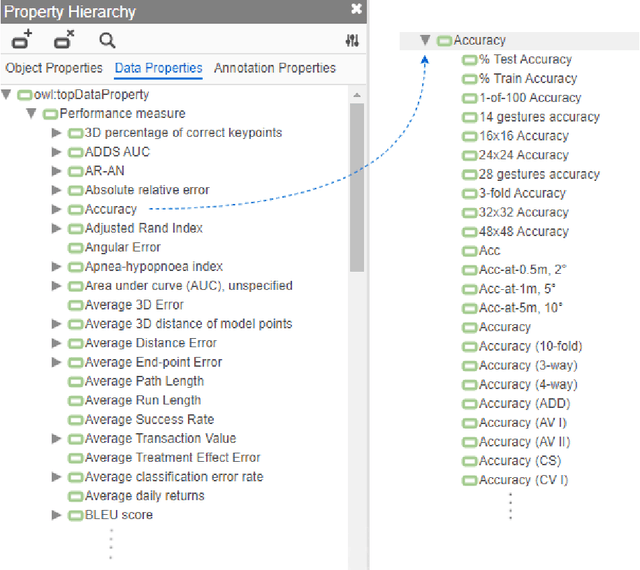
Abstract:Comparing model performances on benchmark datasets is an integral part of measuring and driving progress in artificial intelligence. A model's performance on a benchmark dataset is commonly assessed based on a single or a small set of performance metrics. While this enables quick comparisons, it may also entail the risk of inadequately reflecting model performance if the metric does not sufficiently cover all performance characteristics. Currently, it is unknown to what extent this might impact current benchmarking efforts. To address this question, we analysed the current landscape of performance metrics based on data covering 3867 machine learning model performance results from the web-based open platform 'Papers with Code'. Our results suggest that the large majority of metrics currently used to evaluate classification AI benchmark tasks have properties that may result in an inadequate reflection of a classifiers' performance, especially when used with imbalanced datasets. While alternative metrics that address problematic properties have been proposed, they are currently rarely applied as performance metrics in benchmarking tasks. Finally, we noticed that the reporting of metrics was partly inconsistent and partly unspecific, which may lead to ambiguities when comparing model performances.
Benchmarking neural embeddings for link prediction in knowledge graphs under semantic and structural changes
May 28, 2020



Abstract:Recently, link prediction algorithms based on neural embeddings have gained tremendous popularity in the Semantic Web community, and are extensively used for knowledge graph completion. While algorithmic advances have strongly focused on efficient ways of learning embeddings, fewer attention has been drawn to the different ways their performance and robustness can be evaluated. In this work we propose an open-source evaluation pipeline, which benchmarks the accuracy of neural embeddings in situations where knowledge graphs may experience semantic and structural changes. We define relation-centric connectivity measures that allow us to connect the link prediction capacity to the structure of the knowledge graph. Such an evaluation pipeline is especially important to simulate the accuracy of embeddings for knowledge graphs that are expected to be frequently updated.
Post-hoc explanation of black-box classifiers using confident itemsets
May 05, 2020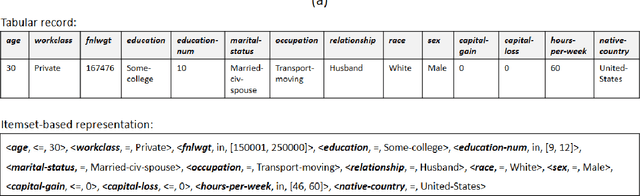
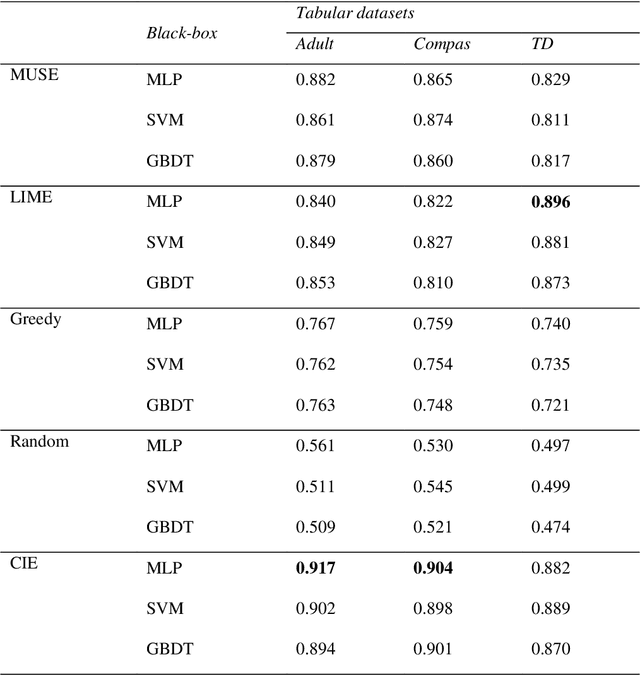
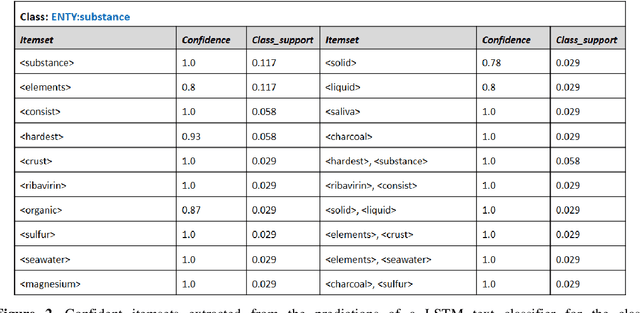
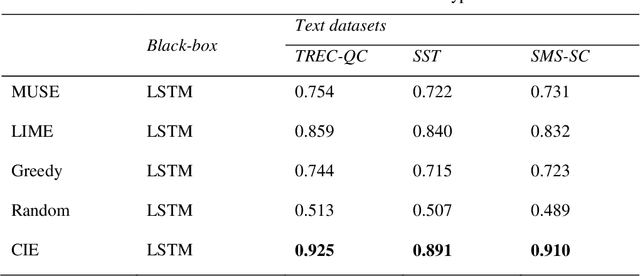
Abstract:It is difficult to trust decisions made by Black-box Artificial Intelligence (AI) methods since their inner working and decision logic is hidden from the user. Explainable Artificial Intelligence (XAI) refers to systems that try to explain how a black-box AI model produces its outcomes. Post-hoc XAI methods approximate the behavior of a black-box by extracting relationships between feature values and the predictions. Some post-hoc explanators randomly perturb data records and build local linear models to explain individual predictions. Other type of explanators use frequent itemsets to extract feature values that frequently appear in samples belonging to a particular class. However, the above methods have some limitations. Random perturbations do not take into account the distribution of feature values in different subspaces, leading to misleading approximations. Frequent itemsets only pay attention to frequently appearing feature values and miss many important correlations between features and class labels that could accurately represent decision boundaries of the model. In this paper, we address the above challenges by proposing an explanation method named Confident Itemsets Explanation (CIE). We introduce confident itemsets, a set of feature values that are highly correlated to a specific class label. CIE utilizes confident itemsets to discretize the whole decision space of a model to smaller subspaces. Extracting important correlations between the features and the outcomes of the black-box in different subspaces, CIE produces instance-wise and class-wise explanations that accurately approximate the behavior of the target black-box classifier.
Dividing the Ontology Alignment Task with Semantic Embeddings and Logic-based Modules
Feb 25, 2020



Abstract:Large ontologies still pose serious challenges to state-of-the-art ontology alignment systems. In this paper we present an approach that combines a neural embedding model and logic-based modules to accurately divide an input ontology matching task into smaller and more tractable matching (sub)tasks. We have conducted a comprehensive evaluation using the datasets of the Ontology Alignment Evaluation Initiative. The results are encouraging and suggest that the proposed method is adequate in practice and can be integrated within the workflow of systems unable to cope with very large ontologies.
OpenBioLink: A resource and benchmarking framework for large-scale biomedical link prediction
Dec 10, 2019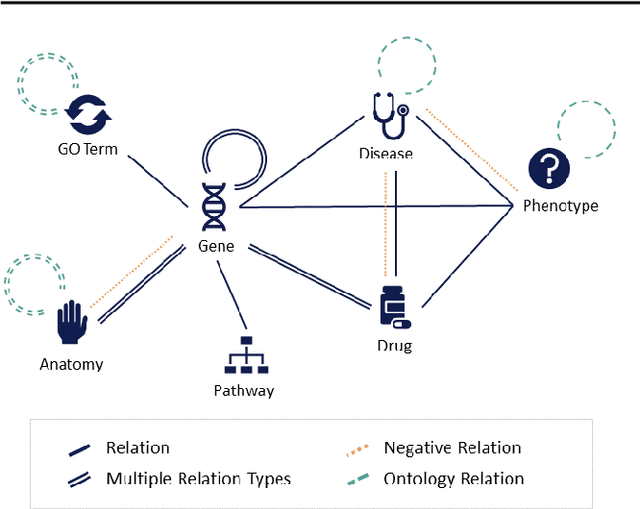
Abstract:SUMMARY: Recently, novel machine-learning algorithms have shown potential for predicting undiscovered links in biomedical knowledge networks. However, dedicated benchmarks for measuring algorithmic progress have not yet emerged. With OpenBioLink, we introduce a large-scale, high-quality and highly challenging biomedical link prediction benchmark to transparently and reproducibly evaluate such algorithms. Furthermore, we present preliminary baseline evaluation results. AVAILABILITY AND IMPLEMENTATION: Source code, data and supplementary files are openly available at https://github.com/OpenBioLink/OpenBioLink CONTACT: matthias.samwald ((at)) meduniwien.ac.at
Clustering of Deep Contextualized Representations for Summarization of Biomedical Texts
Aug 22, 2019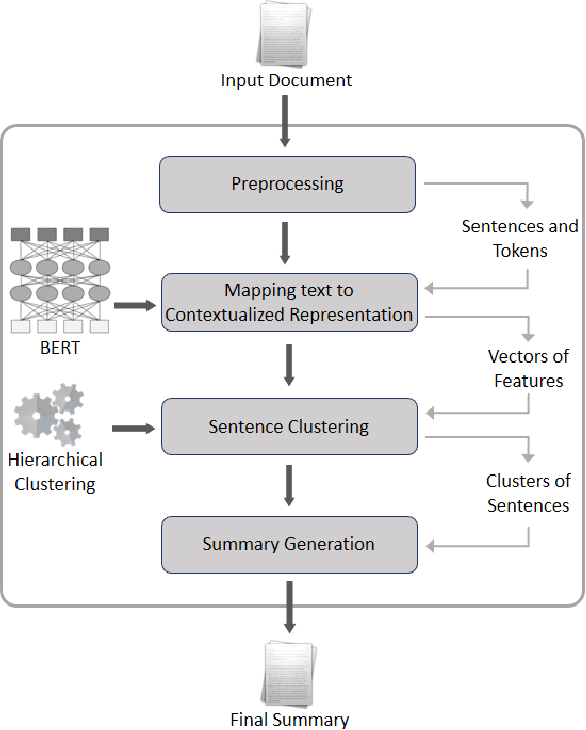
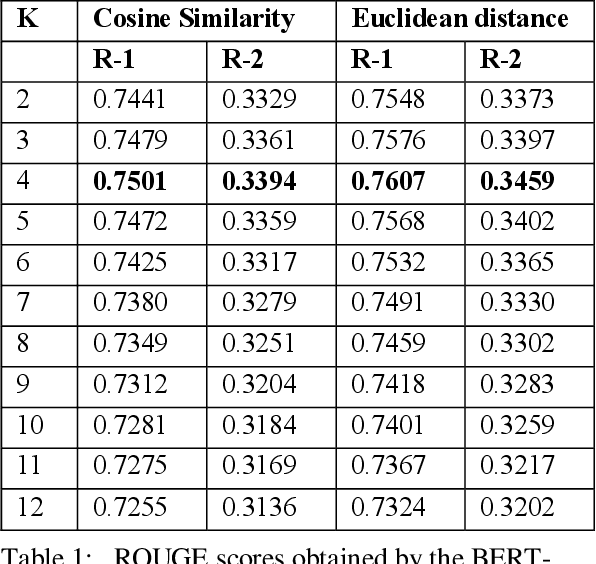
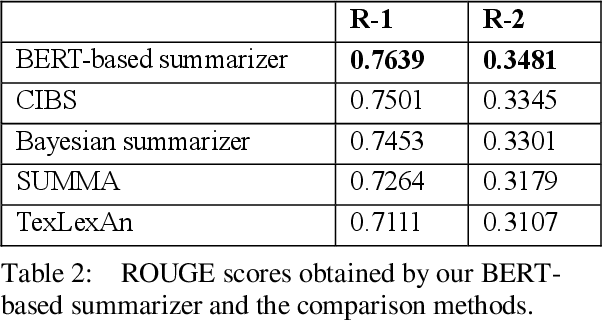
Abstract:In recent years, summarizers that incorporate domain knowledge into the process of text summarization have outperformed generic methods, especially for summarization of biomedical texts. However, construction and maintenance of domain knowledge bases are resource-intense tasks requiring significant manual annotation. In this paper, we demonstrate that contextualized representations extracted from the pre-trained deep language model BERT, can be effectively used to measure the similarity between sentences and to quantify the informative content. The results show that our BERT-based summarizer can improve the performance of biomedical summarization. Although the summarizer does not use any sources of domain knowledge, it can capture the context of sentences more accurately than the comparison methods. The source code and data are available at https://github.com/BioTextSumm/BERT-based-Summ.
Global and local evaluation of link prediction tasks with neural embeddings
Jul 27, 2018



Abstract:We focus our attention on the link prediction problem for knowledge graphs, which is treated herein as a binary classification task on neural embeddings of the entities. By comparing, combining and extending different methodologies for link prediction on graph-based data coming from different domains, we formalize a unified methodology for the quality evaluation benchmark of neural embeddings for knowledge graphs. This benchmark is then used to empirically investigate the potential of training neural embeddings globally for the entire graph, as opposed to the usual way of training embeddings locally for a specific relation. This new way of testing the quality of the embeddings evaluates the performance of binary classifiers for scalable link prediction with limited data. Our evaluation pipeline is made open source, and with this we aim to draw more attention of the community towards an important issue of transparency and reproducibility of the neural embeddings evaluations.
Breaking-down the Ontology Alignment Task with a Lexical Index and Neural Embeddings
May 31, 2018


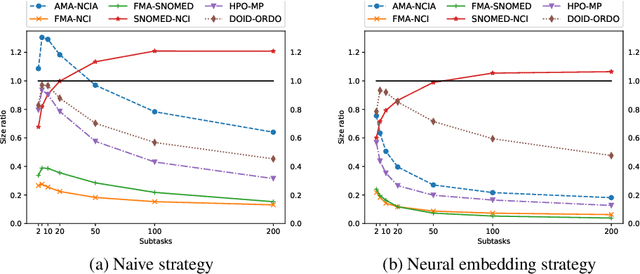
Abstract:Large ontologies still pose serious challenges to state-of-the-art ontology alignment systems. In the paper we present an approach that combines a lexical index, a neural embedding model and locality modules to effectively divide an input ontology matching task into smaller and more tractable matching (sub)tasks. We have conducted a comprehensive evaluation using the datasets of the Ontology Alignment Evaluation Initiative. The results are encouraging and suggest that the proposed methods are adequate in practice and can be integrated within the workflow of state-of-the-art systems.
Fast and scalable learning of neuro-symbolic representations of biomedical knowledge
Apr 30, 2018



Abstract:In this work we address the problem of fast and scalable learning of neuro-symbolic representations for general biological knowledge. Based on a recently published comprehensive biological knowledge graph (Alshahrani, 2017) that was used for demonstrating neuro-symbolic representation learning, we show how to train fast (under 1 minute) log-linear neural embeddings of the entities. We utilize these representations as inputs for machine learning classifiers to enable important tasks such as biological link prediction. Classifiers are trained by concatenating learned entity embeddings to represent entity relations, and training classifiers on the concatenated embeddings to discern true relations from automatically generated negative examples. Our simple embedding methodology greatly improves on classification error compared to previously published state-of-the-art results, yielding a maximum increase of $+0.28$ F-measure and $+0.22$ ROC AUC scores for the most difficult biological link prediction problem. Finally, our embedding approach is orders of magnitude faster to train ($\leq$ 1 minute vs. hours), much more economical in terms of embedding dimensions ($d=50$ vs. $d=512$), and naturally encodes the directionality of the asymmetric biological relations, that can be controlled by the order with which we concatenate the embeddings.
Applying deep learning techniques on medical corpora from the World Wide Web: a prototypical system and evaluation
Feb 12, 2015

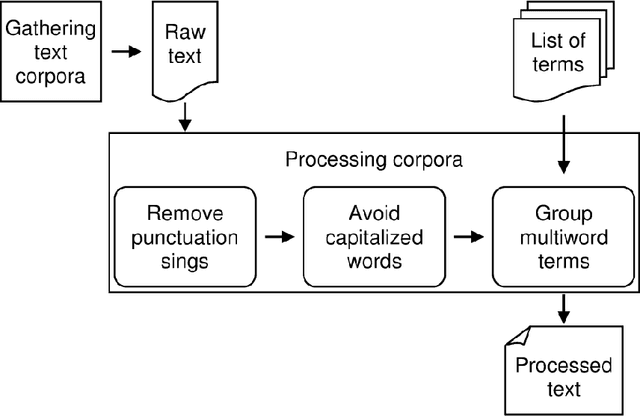
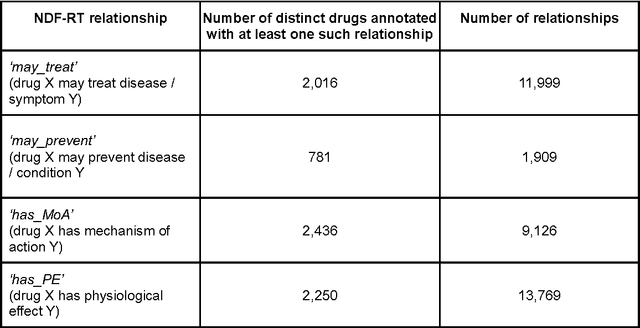
Abstract:BACKGROUND: The amount of biomedical literature is rapidly growing and it is becoming increasingly difficult to keep manually curated knowledge bases and ontologies up-to-date. In this study we applied the word2vec deep learning toolkit to medical corpora to test its potential for identifying relationships from unstructured text. We evaluated the efficiency of word2vec in identifying properties of pharmaceuticals based on mid-sized, unstructured medical text corpora available on the web. Properties included relationships to diseases ('may treat') or physiological processes ('has physiological effect'). We compared the relationships identified by word2vec with manually curated information from the National Drug File - Reference Terminology (NDF-RT) ontology as a gold standard. RESULTS: Our results revealed a maximum accuracy of 49.28% which suggests a limited ability of word2vec to capture linguistic regularities on the collected medical corpora compared with other published results. We were able to document the influence of different parameter settings on result accuracy and found and unexpected trade-off between ranking quality and accuracy. Pre-processing corpora to reduce syntactic variability proved to be a good strategy for increasing the utility of the trained vector models. CONCLUSIONS: Word2vec is a very efficient implementation for computing vector representations and for its ability to identify relationships in textual data without any prior domain knowledge. We found that the ranking and retrieved results generated by word2vec were not of sufficient quality for automatic population of knowledge bases and ontologies, but could serve as a starting point for further manual curation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge