Marina Strocchi
Cardiac Digital Twins at Scale from MRI: Open Tools and Representative Models from ~55000 UK Biobank Participants
May 27, 2025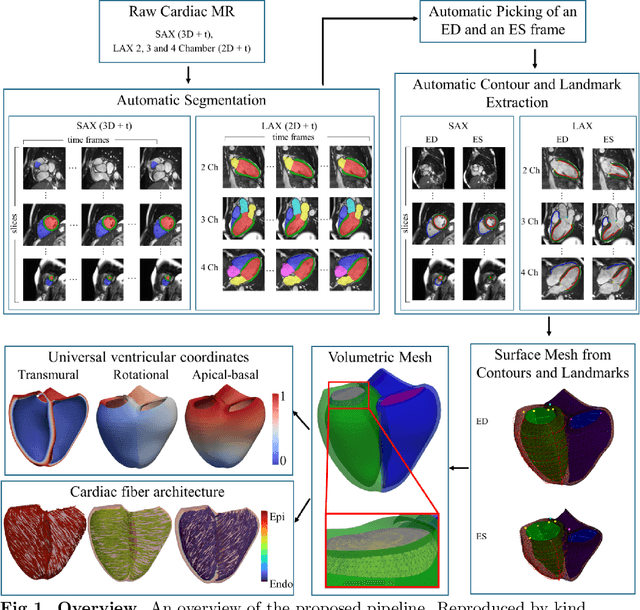
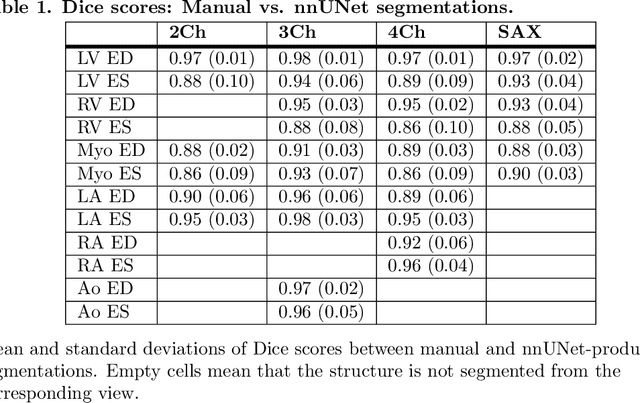
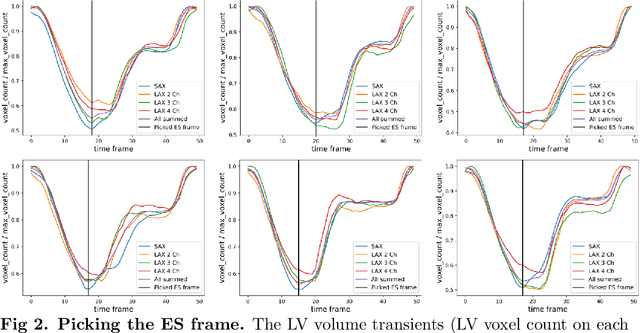
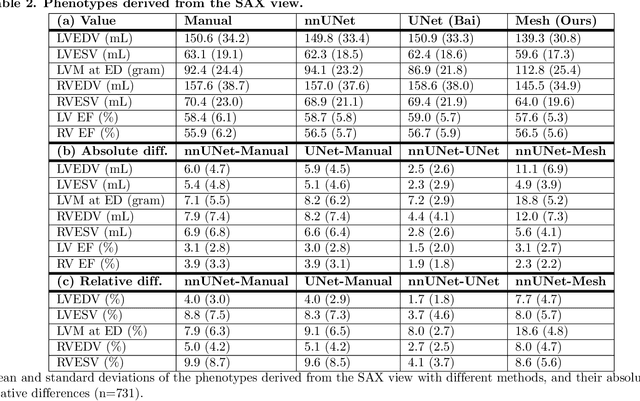
Abstract:A cardiac digital twin is a virtual replica of a patient's heart for screening, diagnosis, prognosis, risk assessment, and treatment planning of cardiovascular diseases. This requires an anatomically accurate patient-specific 3D structural representation of the heart, suitable for electro-mechanical simulations or study of disease mechanisms. However, generation of cardiac digital twins at scale is demanding and there are no public repositories of models across demographic groups. We describe an automatic open-source pipeline for creating patient-specific left and right ventricular meshes from cardiovascular magnetic resonance images, its application to a large cohort of ~55000 participants from UK Biobank, and the construction of the most comprehensive cohort of adult heart models to date, comprising 1423 representative meshes across sex (male, female), body mass index (range: 16 - 42 kg/m$^2$) and age (range: 49 - 80 years). Our code is available at https://github.com/cdttk/biv-volumetric-meshing/tree/plos2025 , and pre-trained networks, representative volumetric meshes with fibers and UVCs will be made available soon.
Real-time whole-heart electromechanical simulations using Latent Neural Ordinary Differential Equations
Jun 08, 2023



Abstract:Cardiac digital twins provide a physics and physiology informed framework to deliver predictive and personalized medicine. However, high-fidelity multi-scale cardiac models remain a barrier to adoption due to their extensive computational costs and the high number of model evaluations needed for patient-specific personalization. Artificial Intelligence-based methods can make the creation of fast and accurate whole-heart digital twins feasible. In this work, we use Latent Neural Ordinary Differential Equations (LNODEs) to learn the temporal pressure-volume dynamics of a heart failure patient. Our surrogate model based on LNODEs is trained from 400 3D-0D whole-heart closed-loop electromechanical simulations while accounting for 43 model parameters, describing single cell through to whole organ and cardiovascular hemodynamics. The trained LNODEs provides a compact and efficient representation of the 3D-0D model in a latent space by means of a feedforward fully-connected Artificial Neural Network that retains 3 hidden layers with 13 neurons per layer and allows for 300x real-time numerical simulations of the cardiac function on a single processor of a standard laptop. This surrogate model is employed to perform global sensitivity analysis and robust parameter estimation with uncertainty quantification in 3 hours of computations, still on a single processor. We match pressure and volume time traces unseen by the LNODEs during the training phase and we calibrate 4 to 11 model parameters while also providing their posterior distribution. This paper introduces the most advanced surrogate model of cardiac function available in the literature and opens new important venues for parameter calibration in cardiac digital twins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge