Lucy L. Gao
Decomposing Gaussians with Unknown Covariance
Sep 17, 2024



Abstract:Common workflows in machine learning and statistics rely on the ability to partition the information in a data set into independent portions. Recent work has shown that this may be possible even when conventional sample splitting is not (e.g., when the number of samples $n=1$, or when observations are not independent and identically distributed). However, the approaches that are currently available to decompose multivariate Gaussian data require knowledge of the covariance matrix. In many important problems (such as in spatial or longitudinal data analysis, and graphical modeling), the covariance matrix may be unknown and even of primary interest. Thus, in this work we develop new approaches to decompose Gaussians with unknown covariance. First, we present a general algorithm that encompasses all previous decomposition approaches for Gaussian data as special cases, and can further handle the case of an unknown covariance. It yields a new and more flexible alternative to sample splitting when $n>1$. When $n=1$, we prove that it is impossible to partition the information in a multivariate Gaussian into independent portions without knowing the covariance matrix. Thus, we use the general algorithm to decompose a single multivariate Gaussian with unknown covariance into dependent parts with tractable conditional distributions, and demonstrate their use for inference and validation. The proposed decomposition strategy extends naturally to Gaussian processes. In simulation and on electroencephalography data, we apply these decompositions to the tasks of model selection and post-selection inference in settings where alternative strategies are unavailable.
Moreau Envelope Based Difference-of-weakly-Convex Reformulation and Algorithm for Bilevel Programs
Jun 29, 2023



Abstract:Recently, Ye et al. (Mathematical Programming 2023) designed an algorithm for solving a specific class of bilevel programs with an emphasis on applications related to hyperparameter selection, utilizing the difference of convex algorithm based on the value function approach reformulation. The proposed algorithm is particularly powerful when the lower level problem is fully convex , such as a support vector machine model or a least absolute shrinkage and selection operator model. In this paper, to suit more applications related to machine learning and statistics, we substantially weaken the underlying assumption from lower level full convexity to weak convexity. Accordingly, we propose a new reformulation using Moreau envelope of the lower level problem and demonstrate that this reformulation is a difference of weakly convex program. Subsequently, we develop a sequentially convergent algorithm for solving this difference of weakly convex program. To evaluate the effectiveness of our approach, we conduct numerical experiments on the bilevel hyperparameter selection problem from elastic net, sparse group lasso, and RBF kernel support vector machine models.
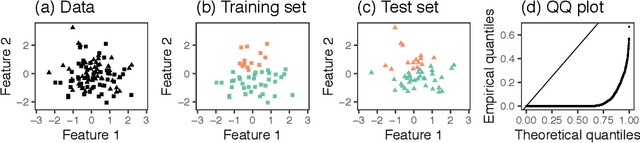
Generalized Data Thinning Using Sufficient Statistics
Mar 22, 2023



Abstract:Our goal is to develop a general strategy to decompose a random variable $X$ into multiple independent random variables, without sacrificing any information about unknown parameters. A recent paper showed that for some well-known natural exponential families, $X$ can be "thinned" into independent random variables $X^{(1)}, \ldots, X^{(K)}$, such that $X = \sum_{k=1}^K X^{(k)}$. In this paper, we generalize their procedure by relaxing this summation requirement and simply asking that some known function of the independent random variables exactly reconstruct $X$. This generalization of the procedure serves two purposes. First, it greatly expands the families of distributions for which thinning can be performed. Second, it unifies sample splitting and data thinning, which on the surface seem to be very different, as applications of the same principle. This shared principle is sufficiency. We use this insight to perform generalized thinning operations for a diverse set of families.
Data thinning for convolution-closed distributions
Jan 18, 2023Abstract:We propose data thinning, a new approach for splitting an observation into two or more independent parts that sum to the original observation, and that follow the same distribution as the original observation, up to a (known) scaling of a parameter. This proposal is very general, and can be applied to any observation drawn from a "convolution closed" distribution, a class that includes the Gaussian, Poisson, negative binomial, Gamma, and binomial distributions, among others. It is similar in spirit to -- but distinct from, and more easily applicable than -- a recent proposal known as data fission. Data thinning has a number of applications to model selection, evaluation, and inference. For instance, cross-validation via data thinning provides an attractive alternative to the "usual" approach of cross-validation via sample splitting, especially in unsupervised settings in which the latter is not applicable. In simulations and in an application to single-cell RNA-sequencing data, we show that data thinning can be used to validate the results of unsupervised learning approaches, such as k-means clustering and principal components analysis.
Tree-Values: selective inference for regression trees
Jun 15, 2021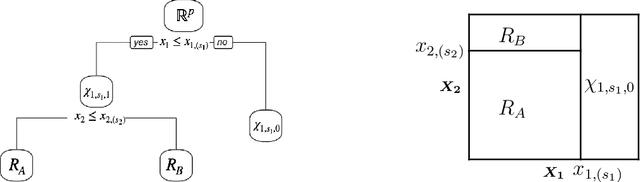
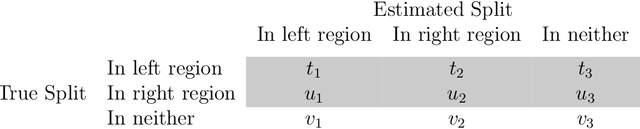
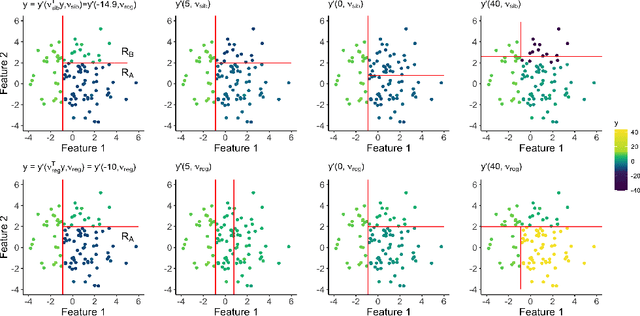
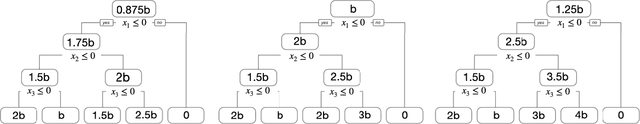
Abstract:We consider conducting inference on the output of the Classification and Regression Tree (CART) [Breiman et al., 1984] algorithm. A naive approach to inference that does not account for the fact that the tree was estimated from the data will not achieve standard guarantees, such as Type 1 error rate control and nominal coverage. Thus, we propose a selective inference framework for conducting inference on a fitted CART tree. In a nutshell, we condition on the fact that the tree was estimated from the data. We propose a test for the difference in the mean response between a pair of terminal nodes that controls the selective Type 1 error rate, and a confidence interval for the mean response within a single terminal node that attains the nominal selective coverage. Efficient algorithms for computing the necessary conditioning sets are provided. We apply these methods in simulation and to a dataset involving the association between portion control interventions and caloric intake.
Selective Inference for Hierarchical Clustering
Dec 05, 2020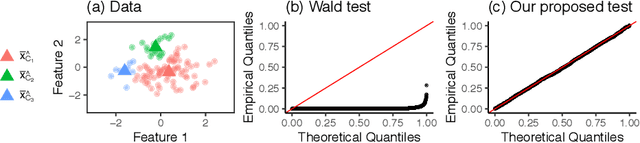

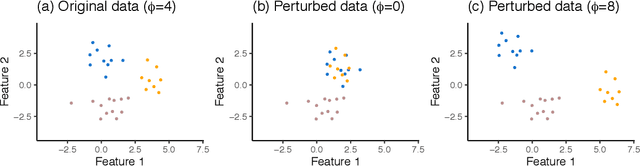
Abstract:Testing for a difference in means between two groups is fundamental to answering research questions across virtually every scientific area. Classical tests control the Type I error rate when the groups are defined a priori. However, when the groups are instead defined via a clustering algorithm, then applying a classical test for a difference in means between the groups yields an extremely inflated Type I error rate. Notably, this problem persists even if two separate and independent data sets are used to define the groups and to test for a difference in their means. To address this problem, in this paper, we propose a selective inference approach to test for a difference in means between two clusters obtained from any clustering method. Our procedure controls the selective Type I error rate by accounting for the fact that the null hypothesis was generated from the data. We describe how to efficiently compute exact p-values for clusters obtained using agglomerative hierarchical clustering with many commonly used linkages. We apply our method to simulated data and to single-cell RNA-seq data.
Testing for Association in Multi-View Network Data
Sep 25, 2019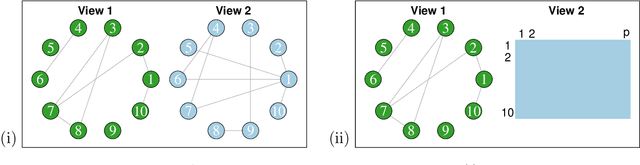
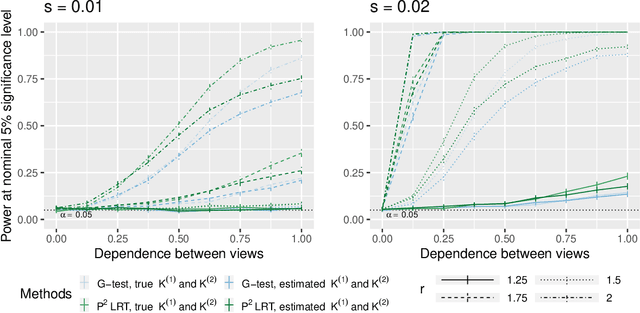
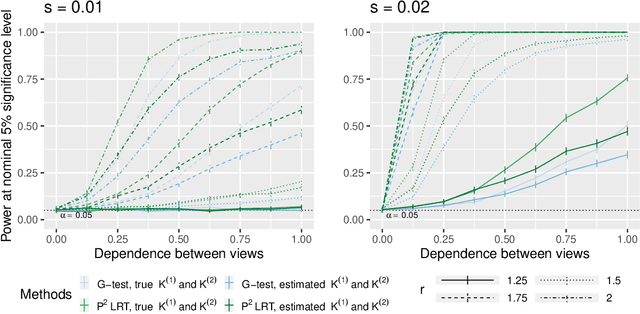
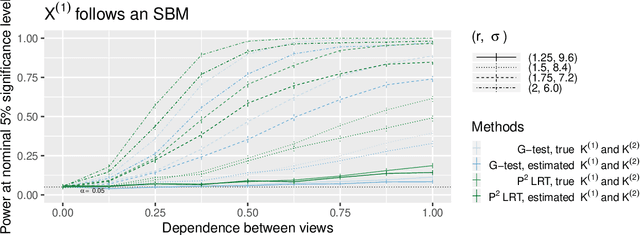
Abstract:In this paper, we consider data consisting of multiple networks, each comprised of a different edge set on a common set of nodes. Many models have been proposed for such multi-view data, assuming that the data views are closely related. In this paper, we provide tools for evaluating the assumption that there is a relationship between the different views. In particular, we ask: is there an association between the latent community memberships of the nodes within each data view? To answer this question, we extend the stochastic block model for a single network view to two network views, and develop a new hypothesis test for the null hypothesis that the latent community structure within each data view is independent. We apply our test to protein-protein interaction data sets from the HINT database (Das & Yu 2012). We find evidence of a weak association between the latent community structure of proteins defined with respect to binary interaction data and with respect to co-complex association data. We also extend this proposal to the setting of a network with node covariates.
Are Clusterings of Multiple Data Views Independent?
Jan 12, 2019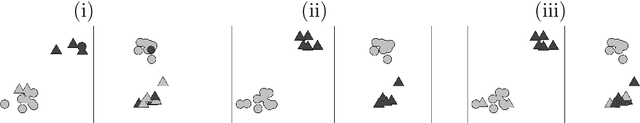
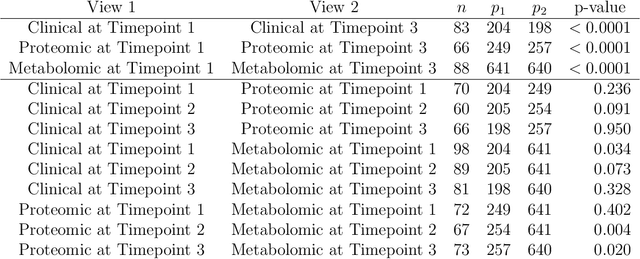
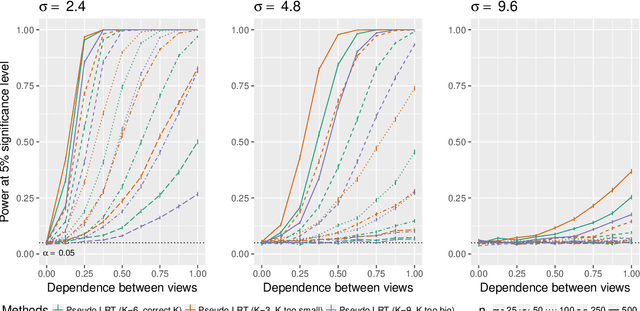
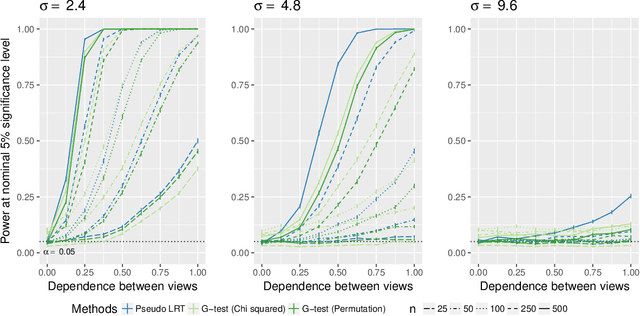
Abstract:In the Pioneer 100 (P100) Wellness Project (Price and others, 2017), multiple types of data are collected on a single set of healthy participants at multiple timepoints in order to characterize and optimize wellness. One way to do this is to identify clusters, or subgroups, among the participants, and then to tailor personalized health recommendations to each subgroup. It is tempting to cluster the participants using all of the data types and timepoints, in order to fully exploit the available information. However, clustering the participants based on multiple data views implicitly assumes that a single underlying clustering of the participants is shared across all data views. If this assumption does not hold, then clustering the participants using multiple data views may lead to spurious results. In this paper, we seek to evaluate the assumption that there is some underlying relationship among the clusterings from the different data views, by asking the question: are the clusters within each data view dependent or independent? We develop a new test for answering this question, which we then apply to clinical, proteomic, and metabolomic data, across two distinct timepoints, from the P100 study. We find that while the subgroups of the participants defined with respect to any single data type seem to be dependent across time, the clustering among the participants based on one data type (e.g. proteomic data) appears not to be associated with the clustering based on another data type (e.g. clinical data).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge