Louis Scheffer
Janelia Farm Research Campus- HHMI
Autoproof: Automated Segmentation Proofreading for Connectomics
Sep 30, 2025Abstract:Producing connectomes from electron microscopy (EM) images has historically required a great deal of human proofreading effort. This manual annotation cost is the current bottleneck in scaling EM connectomics, for example, in making larger connectome reconstructions feasible, or in enabling comparative connectomics where multiple related reconstructions are produced. In this work, we propose using the available ground-truth data generated by this manual annotation effort to learn a machine learning model to automate or optimize parts of the required proofreading workflows. We validate our approach on a recent complete reconstruction of the \emph{Drosophila} male central nervous system. We first show our method would allow for obtaining 90\% of the value of a guided proofreading workflow while reducing required cost by 80\%. We then demonstrate a second application for automatically merging many segmentation fragments to proofread neurons. Our system is able to automatically attach 200 thousand fragments, equivalent to four proofreader years of manual work, and increasing the connectivity completion rate of the connectome by 1.3\% points.
Small Sample Learning of Superpixel Classifiers for EM Segmentation- Extended Version
Jun 13, 2014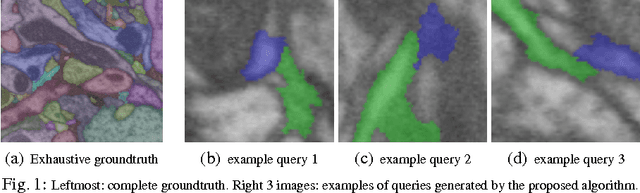
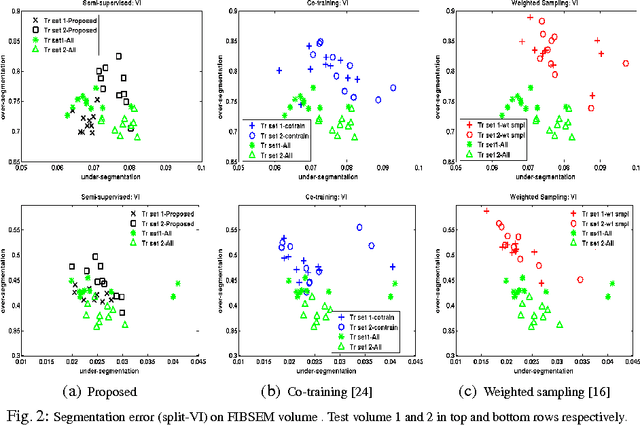
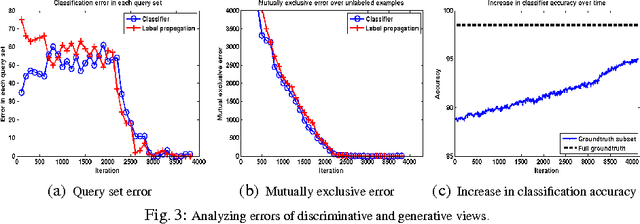
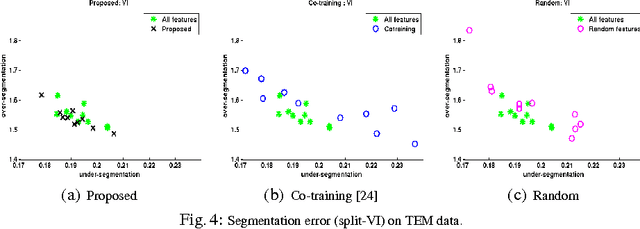
Abstract:Pixel and superpixel classifiers have become essential tools for EM segmentation algorithms. Training these classifiers remains a major bottleneck primarily due to the requirement of completely annotating the dataset which is tedious, error-prone and costly. In this paper, we propose an interactive learning scheme for the superpixel classifier for EM segmentation. Our algorithm is "active semi-supervised" because it requests the labels of a small number of examples from user and applies label propagation technique to generate these queries. Using only a small set ($<20\%$) of all datapoints, the proposed algorithm consistently generates a classifier almost as accurate as that estimated from a complete groundtruth. We provide segmentation results on multiple datasets to show the strength of these classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge